1VJO
 
 | |
1VR3
 
 | |
1VRM
 
 | |
1VQ0
 
 | |
1VJ1
 
 | |
1VPZ
 
 | |
1VQR
 
 | |
1VR8
 
 | |
1VK3
 
 | |
1VL4
 
 | |
1VKH
 
 | |
1ZCZ
 
 | |
1ZKG
 
 | |
1Z9F
 
 | |
1ZX8
 
 | |
1HTM
 
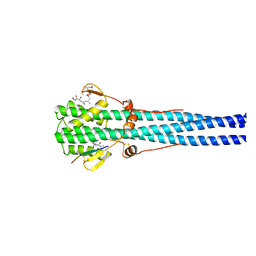 | | STRUCTURE OF INFLUENZA HAEMAGGLUTININ AT THE PH OF MEMBRANE FUSION | | Descriptor: | HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN | | Authors: | Bullough, P.A, Hughson, F.M, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1994-11-02 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of influenza haemagglutinin at the pH of membrane fusion.
Nature, 371, 1994
|
|
2GVI
 
 | |
2IAY
 
 | |
2ICH
 
 | |
2IIZ
 
 | |
2GAZ
 
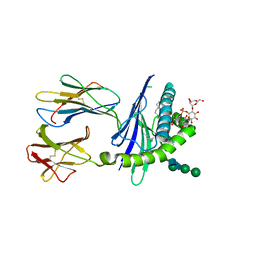 | | Mycobacterial lipoglycan presentation by CD1d | | Descriptor: | (2R)-3-[(HYDROXY{[(2R,3R,5S,6R)-3,4,5-TRIHYDROXY-2,6-BIS(ALPHA-D-MANNOPYRANOSYLOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]PROPAN E-1,2-DIYL DIHEXADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-cell surface glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2006-03-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d.
J.Immunol., 177, 2006
|
|
2FNO
 
 | |
2HAG
 
 | |
2FO4
 
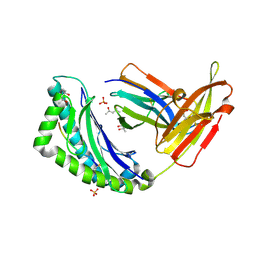 | |
2H1T
 
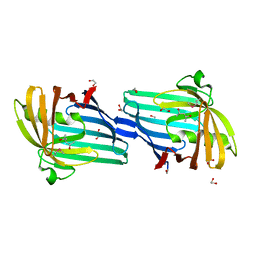 | |
