8I4W
 
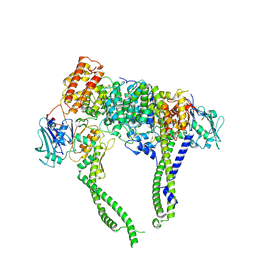 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4X
 
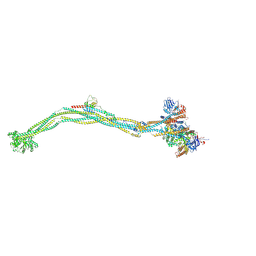 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6K13
 
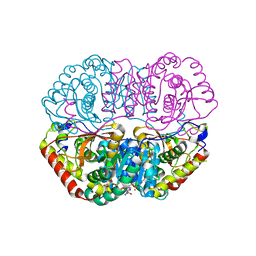 | | Crystal Structure Basis for BmLDH Complex | | Descriptor: | L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID | | Authors: | Long, Y, Shen, Z. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
5DZN
 
 | | human T-cell immunoglobulin and mucin domain protein 4 | | Descriptor: | T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Yuan, S, Rao, Z, Wang, X. | | Deposit date: | 2015-09-25 | | Release date: | 2015-11-25 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope.
Protein Cell, 6, 2015
|
|
6K12
 
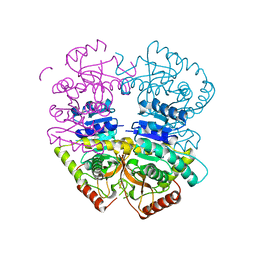 | | Babesia microti lactate dehydrogenase apo form (BmLDH) | | Descriptor: | L-lactate dehydrogenase | | Authors: | Long, Y. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.99 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3R
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.36 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.52 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5D7C
 
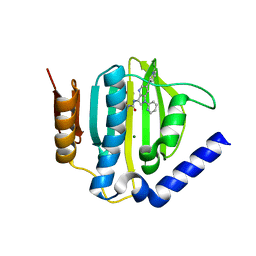 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
3W95
 
 | |
7YQH
 
 | | Cryo-EM structure of 8-subunit Smc5/6 | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 3, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2022-08-07 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
6JZZ
 
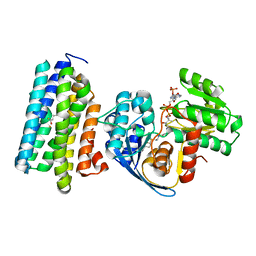 | | The crystal structure of AAR-C294S in complex with ADO. | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZY
 
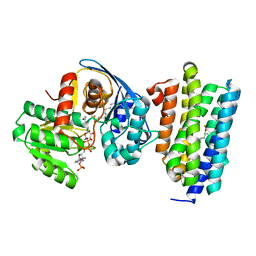 | | Crystal structure of AAR with NADPH and stearyl in complex with ADO binding a long chain carbohydrate | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZU
 
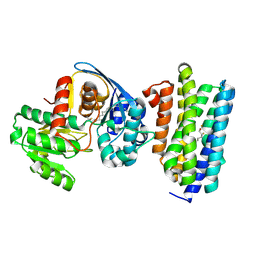 | | The crystal structure of acyl-acyl carrier protein (acyl-ACP) reductase (AAR) in complex with aldehyde deformylating oxygenase (ADO) | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZQ
 
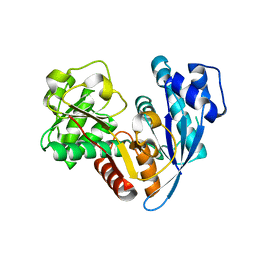 | |
7VH8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Zhao, Y, Zhang, Q, Yang, H, Rao, Z. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332.
Protein Cell, 13, 2022
|
|
5VEB
 
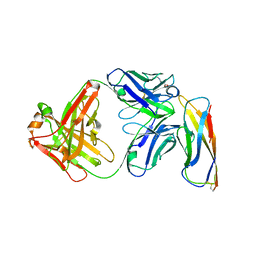 | | Crystal structure of a Fab binding to extracellular domain 5 of Cadherin-6 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Cadherin-6, anti-CDH6 Fab heavy chain, ... | | Authors: | Zhu, X, Bialucha, C.U, London, A, Clark, K, Hu, T. | | Deposit date: | 2017-04-04 | | Release date: | 2017-06-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Optimization of HKT288, a Cadherin-6-Targeting ADC for the Treatment of Ovarian and Renal Cancers.
Cancer Discov, 7, 2017
|
|
4RTI
 
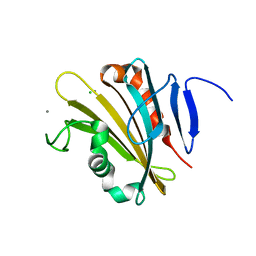 | | The crystal structure of PsbP from Spinacia oleracea | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Oxygen-evolving enhancer protein 2, ... | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
4RTH
 
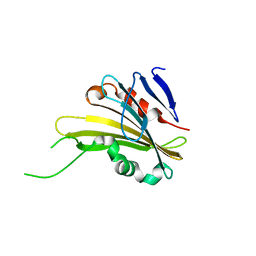 | | The crystal structure of PsbP from Zea mays | | Descriptor: | Membrane-extrinsic protein of photosystem II PsbP | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
3PL9
 
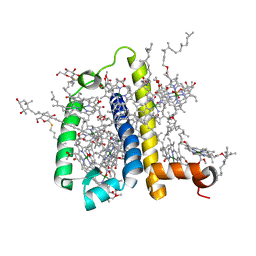 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | Deposit date: | 2010-11-14 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3CPC
 
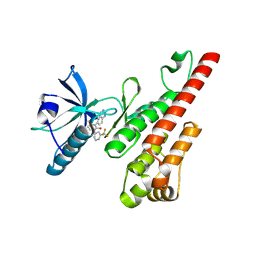 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridone inhibitor | | Descriptor: | 3-(2-aminoquinazolin-6-yl)-4-methyl-1-[3-(trifluoromethyl)phenyl]pyridin-2(1H)-one, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-03-31 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Aryl Aminoquinazoline Pyridones as Potent, Selective, and Orally Efficacious Inhibitors of Receptor Tyrosine Kinase c-Kit.
J.Med.Chem., 51, 2008
|
|
1Q3H
 
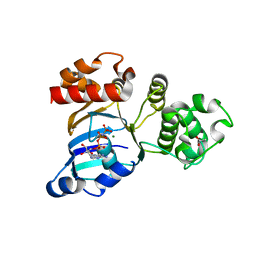 | | mouse CFTR NBD1 with AMP.PNP | | Descriptor: | ACETIC ACID, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
