6BRD
 
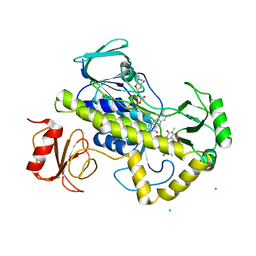 | | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complexed with rifampin and FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Cox, G, Kelso, J, Stogios, P.J, Savchenko, A, Anderson, W.F, Wright, G.D, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-30 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Cell Chem Biol, 25, 2018
|
|
3V4E
 
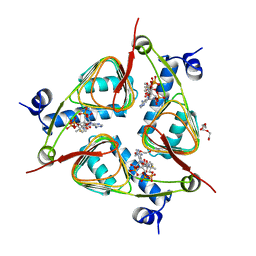 | | Crystal Structure of the galactoside O-acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, Galactoside O-acetyltransferase, ... | | Authors: | Knapik, A.A, Shumilin, I.A, Luo, H.-B, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
1L61
 
 | |
6D7Y
 
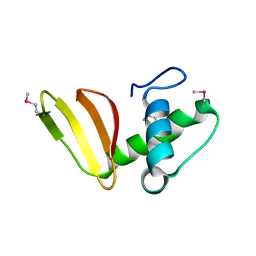 | | 1.75 Angstrom Resolution Crystal Structure of the Toxic C-Terminal Tip of CdiA from Pseudomonas aeruginosa in Complex with Immune Protein | | Descriptor: | Hemagglutinin, immune protein | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Allen, J.P, Hauser, A.R, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-25 | | Release date: | 2019-05-01 | | Last modified: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A comparative genomics approach identifies contact-dependent growth inhibition as a virulence determinant.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3TZC
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(Y155F) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Domagalski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
6CTY
 
 | | Crystal structure of dihydroorotase pyrC from Yersinia pestis in complex with zinc and malate at 2.4 A resolution | | Descriptor: | D-MALATE, Dihydroorotase, ZINC ION | | Authors: | Lipowska, J, Shabalin, I.G, Winsor, J, Woinska, M, Cooper, D.R, Kwon, K, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-23 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
3TZH
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(F187A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, GLYCEROL, SULFATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Cooper, D.R, Osinski, T, Shumilin, I, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: |
|
|
3U09
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(G92D) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Fratczak, Z, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3TYK
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | Descriptor: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
1L42
 
 | |
1L70
 
 | |
1L75
 
 | |
4R6I
 
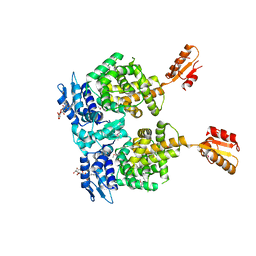 | | AtxA protein, a virulence regulator from Bacillus anthracis. | | Descriptor: | Anthrax toxin expression trans-acting positive regulator, DODECYL-BETA-D-MALTOSIDE | | Authors: | Osipiuk, J, Horton, L.B, Koehler, T.M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-10-22 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity.
Mol.Microbiol., 95, 2015
|
|
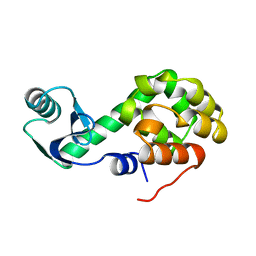
1L47
 
 | |
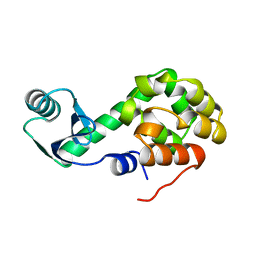
1L50
 
 | |
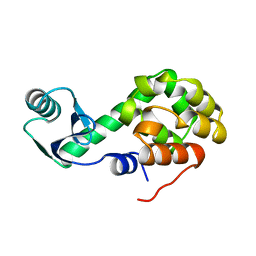
1L45
 
 | |
1L69
 
 | |
1L48
 
 | |
4RV8
 
 | | Co-Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum and the inhibitor p131 | | Descriptor: | 1-(2-{3-[(1E)-N-(2-aminoethoxy)ethanimidoyl]phenyl}propan-2-yl)-3-(4-chloro-3-nitrophenyl)urea, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-25 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structure of Cryptosporidium IMP dehydrogenase bound to an inhibitor with in vivo antiparasitic activity.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
1L51
 
 | |
1L56
 
 | |
1L60
 
 | |
1L67
 
 | |
1L74
 
 | |
1L73
 
 | |
