3OF6
 
 | | Human pre-T cell receptor crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre T-cell antigen receptor alpha, T cell receptor beta chain | | Authors: | Pang, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Nature, 467, 2010
|
|
4PJ5
 
 | | Structure of human MR1-Ac-6-FP in complex with human MAIT TRBV6-1 TCR | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
4PJI
 
 | |
4PRN
 
 | | Crystal structure of a HLA-B*35:01-HPVG-A4 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRD
 
 | | Crystal structure of a HLA-B*35:08-HPVG-D5 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
7WK0
 
 | | Local refine of Omicron spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
4PGF
 
 | | The structure of mono-acetylated SAHH | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kavran, J.M, Wang, Y, Cole, P.A, Leahy, D.J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Regulation of s-adenosylhomocysteine hydrolase by lysine acetylation.
J.Biol.Chem., 289, 2014
|
|
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
4PJD
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT C-C10 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
7WJZ
 
 | | Omicron Spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
4PJC
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT C-A11 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
7WOB
 
 | | SARS-CoV-2 Spike in complex with IgG 553-60 (2-up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOG
 
 | | SARS-CoV-2 Omicron S monomer complexed with 553-49 | | Descriptor: | 553-49 VH, 553-49 VL, Spike protein S1 | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO4
 
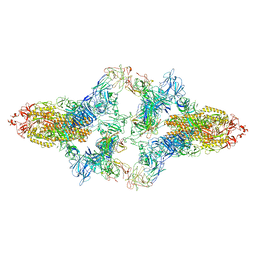 | | SARS-CoV-2 Spike in complex with IgG 553-15 (S-553-15 dimer trimer ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO5
 
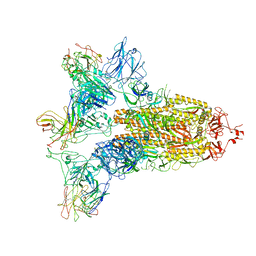 | | SARS-CoV-2 Spike in complex with IgG 553-15 (S-553-15 trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO7
 
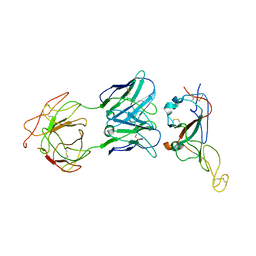 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOC
 
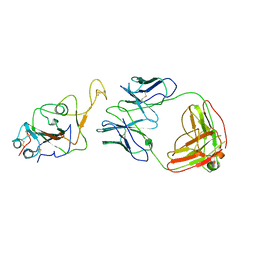 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOA
 
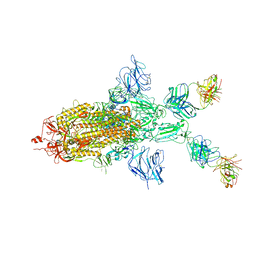 | | SARS-CoV-2 Spike in complex with IgG 553-60 (1-up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
4PRE
 
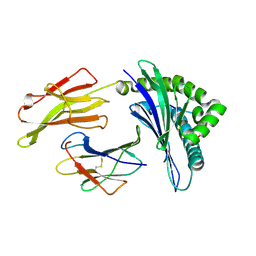 | | Crystal structure of a HLA-B*35:08-HPVG-Q5 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRB
 
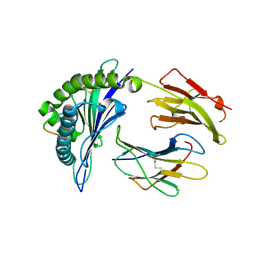 | | Crystal structure of a HLA-B*35:08-HPVG-A4 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PJE
 
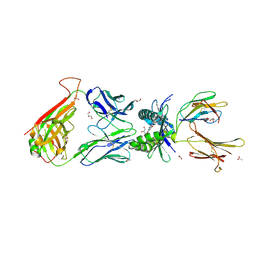 | |
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
4PJ8
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT TRBV20 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
4PJF
 
 | |
