7LDQ
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2846 | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-13 | | Release date: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2846
To Be Published
|
|
7LGP
 
 | | DapE enzyme from Shigella flexneri | | Descriptor: | CHLORIDE ION, SODIUM ION, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Osipiuk, J, Endres, M, Becker, D.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DapE enzyme from Shigella flexneri
To Be Published
|
|
1V4A
 
 | | Structure of the N-terminal Domain of Escherichia coli Glutamine Synthetase adenylyltransferase | | Descriptor: | Glutamate-ammonia-ligase adenylyltransferase | | Authors: | Xu, Y, Zhang, R, Joachimiak, A, Carr, P.D, Ollis, D.L, Vasudevan, S.G. | | Deposit date: | 2003-11-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the n-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase
Structure, 12, 2004
|
|
2VB1
 
 | | HEWL at 0.65 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, LYSOZYME C, ... | | Authors: | Wang, J, Dauter, M, Alkire, R, Joachimiak, A, Dauter, Z. | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (0.65 Å) | | Cite: | Triclinic Lysozyme at 0.65 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4K0D
 
 | | Periplasmic sensor domain of sensor histidine kinase, Adeh_2942 | | Descriptor: | ACETATE ION, CHLORIDE ION, Periplasmic sensor hybrid histidine kinase, ... | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-12 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7KYU
 
 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
2A5L
 
 | | The crystal structure of the Trp repressor binding protein WrbA from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, Trp repressor binding protein WrbA | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the Trp repressor binding protein WrbA from Pseudomonas aeruginosa
To be Published
|
|
2RE1
 
 | | Crystal structure of aspartokinase alpha and beta subunits | | Descriptor: | Aspartokinase, alpha and beta subunits, CALCIUM ION | | Authors: | Chang, C, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of aspartokinase alpha and beta subunits.
To be Published
|
|
2QSX
 
 | | Crystal structure of putative transcriptional regulator LysR From Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator, LysR family, SULFATE ION | | Authors: | Wu, R, Abdullah, J, Binkowski, T.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Putative Transcriptional Regulator LysR From Vibrio parahaemolyticus.
To be Published
|
|
1T06
 
 | |
1T0T
 
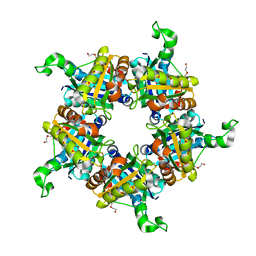 | | Crystallographic structure of a putative chlorite dismutase | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, APC35880, MAGNESIUM ION | | Authors: | Gilski, M, Borek, D, Chen, Y, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of APC35880 protein from Bacillus Stearothermophilus
To be Published
|
|
1Z7A
 
 | | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-24 | | Release date: | 2005-05-10 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1
To be Published
|
|
1SRV
 
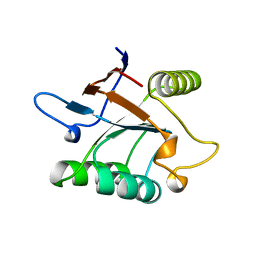 | | THERMUS THERMOPHILUS GROEL (HSP60 CLASS) FRAGMENT (APICAL DOMAIN) COMPRISING RESIDUES 192-336 | | Descriptor: | PROTEIN (GROEL (HSP60 CLASS)) | | Authors: | Walsh, M.A, Dementieva, I, Evans, G, Sanishvili, R, Joachimiak, A. | | Deposit date: | 1999-03-02 | | Release date: | 1999-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Taking MAD to the extreme: ultrafast protein structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1T6A
 
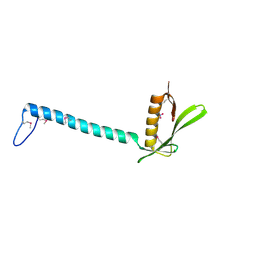 | | Crystal Structure of Protein of Unknown Function from Bacillus stearothermophilus | | Descriptor: | NITRATE ION, RBSTP2229 gene product | | Authors: | Osipiuk, J, Wu, R, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of hypothetical protein (RBSTP2229 gene product) from Bacillus stearothermophilus
To be Published
|
|
1T6T
 
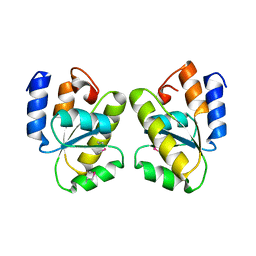 | |
1X87
 
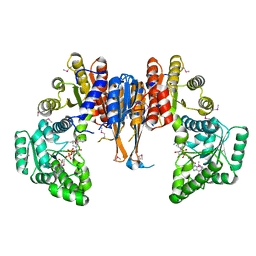 | | 2.4A X-ray structure of Urocanase protein complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Urocanase protein | | Authors: | Tereshko, V, Zaborske, J, Gilbreth, R, Dementieva, I, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-17 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A X-ray structure of Urocanase protein complexed with NAD
To be Published
|
|
1U61
 
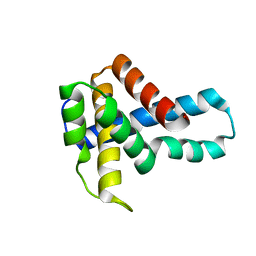 | | Crystal Structure of Putative Ribonuclease III from Bacillus cereus | | Descriptor: | hypothetical protein | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystal structure of conserved hypothetical protein from Bacillus cereus
To be Published
|
|
1T33
 
 | |
1NC5
 
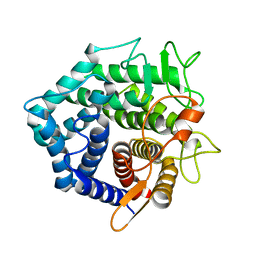 | | Structure of Protein of Unknown Function of YteR from Bacillus Subtilis | | Descriptor: | hypothetical protein yTER | | Authors: | Zhang, R, Lozondra, L, Korolev, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6 A crystal structure of YteR protein from Bacillus subtilis, a predicted lyase.
Proteins, 60, 2005
|
|
1NPY
 
 | | Structure of shikimate 5-dehydrogenase-like protein HI0607 | | Descriptor: | ACETYL GROUP, Hypothetical shikimate 5-dehydrogenase-like protein HI0607 | | Authors: | Korolev, S, Koroleva, O, Zarembinski, T, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a Novel Shikimate Dehydrogenase from Haemophilus influenzae.
J.Biol.Chem., 280, 2005
|
|
4HTQ
 
 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 6.70 x 10e+11 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.Z, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1PC6
 
 | | Structural Genomics, NinB | | Descriptor: | BETA-MERCAPTOETHANOL, Protein ninB | | Authors: | Zhang, R, Beasley, S, Maxwell, K.L, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-15 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Functional similarities between phage lambda Orf and Escherichia coli RecFOR in initiation of genetic exchange
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1K7J
 
 | | Structural Genomics, protein TF1 | | Descriptor: | Protein yciO, SULFATE ION | | Authors: | Zhang, R, Dementieva, I, Thorn, J, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Genomics, protein TF1
To be Published
|
|
