4XGA
 
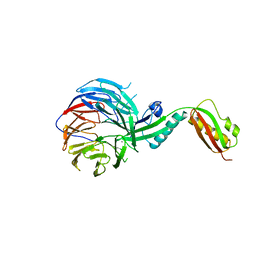 | | Crystal structure of BamB and BamA P3-5 complex from E.coli | | Descriptor: | CALCIUM ION, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB | | Authors: | Chen, Z, Zhan, L.H, Dong, C, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2014-12-30 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the interaction of BamB with the POTRA3-4 domains of BamA.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4E8B
 
 | |
4E0Q
 
 | |
4EQ6
 
 | |
4F1R
 
 | |
4DCM
 
 | |
3Q6B
 
 | |
3TKA
 
 | | crystal structure and solution saxs of methyltransferase rsmh from E.coli | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Ribosomal RNA small subunit methyltransferase H, S-ADENOSYLMETHIONINE, ... | | Authors: | Gao, Z.Q, Wei, Y, Zhang, H, Dong, Y.H. | | Deposit date: | 2011-08-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA.
J.Struct.Biol., 179, 2012
|
|
7Y8M
 
 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, reductase | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7CGS
 
 | |
6JDP
 
 | |
7Y8L
 
 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(2,5-difluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 5-[2,5-bis(fluoranyl)phenyl]-3,4-dihydro-2~{H}-pyrrole, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7Y8K
 
 | | Structure of ScIRED wild-type from Streptomyces clavuligerus | | Descriptor: | Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7V4X
 
 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V50
 
 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V51
 
 | | BVMO_negative mutant D432V | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H.-L. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7Y8N
 
 | | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(2,5-difluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 5-[2,5-bis(fluoranyl)phenyl]-3,4-dihydro-2~{H}-pyrrole, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7Y8O
 
 | | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SciR | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole
to be published
|
|
