6KYW
 
 | | S8-mSRK-S8-SP11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor protein kinase SRK8, S locus protein 11 | | Authors: | Murase, K, Hakoshima, T, Mori, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (2.60112739 Å) | | Cite: | Mechanism of self/nonself-discrimination in Brassica self-incompatibility.
Nat Commun, 11, 2020
|
|
1IQW
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MOUSE ANTI-HUMAN FAS ANTIBODY HFE7A | | Descriptor: | ANTIBODY M-HFE7A, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Ito, S, Takayama, T, Hanzawa, H, Ichikawa, K, Ohsumi, J, Serizawa, N, Hata, T, Haruyama, H. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the antigen-binding fragment of apoptosis-inducing mouse anti-human Fas monoclonal antibody HFE7A.
J.Biochem., 131, 2002
|
|
6YPE
 
 | |
6JEO
 
 | | Structure of PSI tetramer from Anabaena | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-02-06 | | Release date: | 2019-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
5Z8Y
 
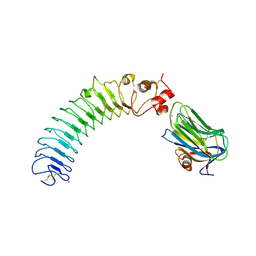 | | Crystal structure of human LRRTM2 in complex with Neurexin 1beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2018-02-01 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into modulation and selectivity of transsynaptic neurexin-LRRTM interaction.
Nat Commun, 9, 2018
|
|
5Z8X
 
 | | Crystal structure of human LRRTM2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat transmembrane neuronal protein 2 | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2018-02-01 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into modulation and selectivity of transsynaptic neurexin-LRRTM interaction.
Nat Commun, 9, 2018
|
|
1V77
 
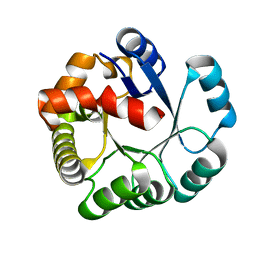 | | Crystal structure of the PH1877 protein | | Descriptor: | hypothetical protein PH1877 | | Authors: | Takagi, H, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 319, 2004
|
|
7WJT
 
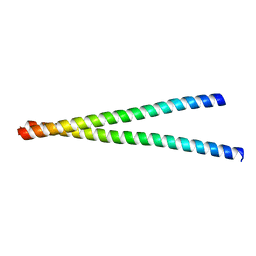 | |
5GZ6
 
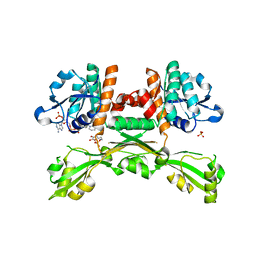 | | Structure of D-amino acid dehydrogenase in complex with NADPH and 2-keto-6-aminocapronic acid | | Descriptor: | 6-azanyl-2-oxidanylidene-hexanoic acid, ACETATE ION, Meso-diaminopimelate D-dehydrogenase, ... | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ3
 
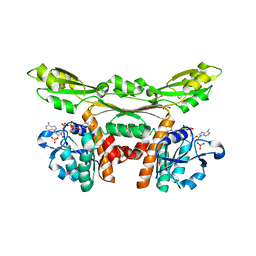 | | Structure of D-amino acid dehydrogenase in complex with NADP | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ1
 
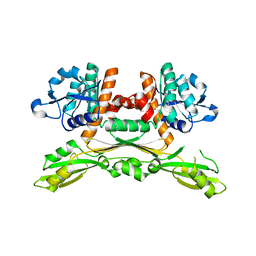 | | Structure of substrate/cofactor-free D-amino acid dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
2Z2E
 
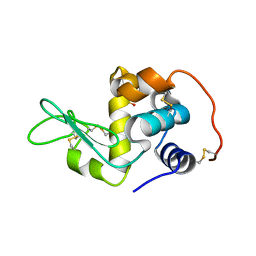 | | Crystal Structure of Canine Milk Lysozyme Stabilized against Non-enzymatic Deamidation | | Descriptor: | Lysozyme C, milk isozyme, SULFATE ION | | Authors: | Nonaka, Y, Akieda, D, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Spontaneous asparaginyl deamidation of canine milk lysozyme under mild conditions
Proteins, 72, 2008
|
|
3VTM
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING INDIUM | | Authors: | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | Deposit date: | 2012-05-31 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective binding of antimicrobial porphyrins to the heme-receptor IsdH-NEAT3 of Staphylococcus aureus
Protein Sci., 22, 2013
|
|
3VUA
 
 | | Apo IsdH-NEAT3 in space group P3121 at a resolution of 1.85 A | | Descriptor: | ACETATE ION, GLYCEROL, Iron-regulated surface determinant protein H, ... | | Authors: | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | Deposit date: | 2012-06-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin
To be Published
|
|
7CE4
 
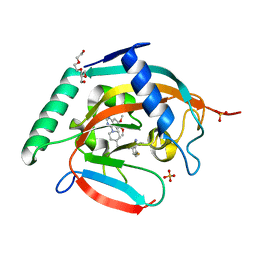 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
3ST7
 
 | | Crystal Structure of capsular polysaccharide assembling protein CapF from staphylococcus aureus | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, GLYCEROL, ZINC ION | | Authors: | Miyafusa, T, Tanaka, Y, Kuroda, M, Yao, M, Watanabe, M, Ohta, T, Tanaka, I, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-07-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains.
Biochem.J., 443, 2012
|
|
