9MDZ
 
 | | anti-IL23R VHH | | Descriptor: | GLYCEROL, anti-IL23R VHH | | Authors: | Kiefer, J.R, Koerber, J.T, Ota, N, Davies, C, Wallweber, H.A. | | Deposit date: | 2024-12-05 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Engineering a protease-stable, oral single-domain antibody to inhibit IL-23 signaling.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9MFG
 
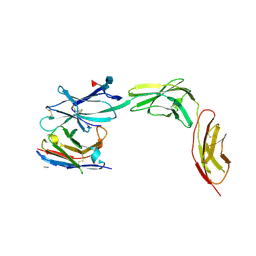 | | Complex of IL23 receptor and VHH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Wallweber, H.A, Koerber, J.T, Ota, N, Davies, C. | | Deposit date: | 2024-12-09 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Engineering a protease-stable, oral single-domain antibody to inhibit IL-23 signaling.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9J8V
 
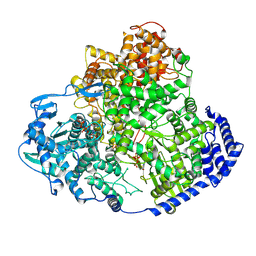 | | TSWV L protein in complex with ribavirin 5-triphosphate | | Descriptor: | RIBAVIRIN TRIPHOSPHATE, RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2024-08-21 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
4DXW
 
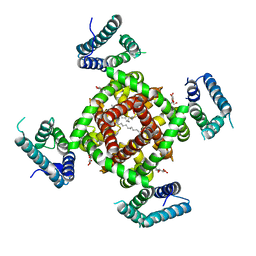 | | Crystal structure of NavRh, a voltage-gated sodium channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Zhang, X, Ren, W.L, Yan, C.Y, Wang, J.W, Yan, N. | | Deposit date: | 2012-02-28 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel
Nature, 486, 2012
|
|
3LUT
 
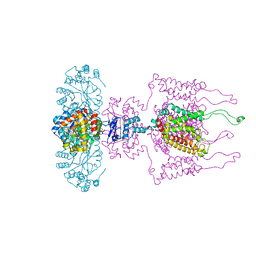 | | A Structural Model for the Full-length Shaker Potassium Channel Kv1.2 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Chen, X, Ni, F, Wang, Q, Ma, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8WO7
 
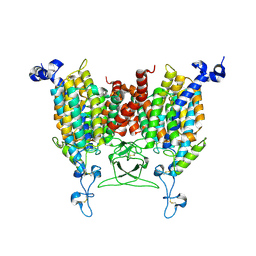 | | Apo state of Arabidopsis AZG1 T440Y | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-10 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8WMQ
 
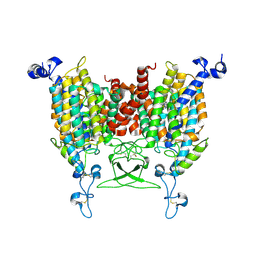 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH5.5 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
2TDX
 
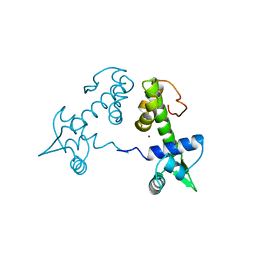 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT) COMPLEXED WITH NICKEL | | Descriptor: | DIPHTHERIA TOX REPRESSOR, NICKEL (II) ION | | Authors: | White, A, Ding, X, Zheng, H, Schiering, N, Ringe, D, Murphy, J.R. | | Deposit date: | 1998-06-22 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex.
Nature, 394, 1998
|
|
7BV5
 
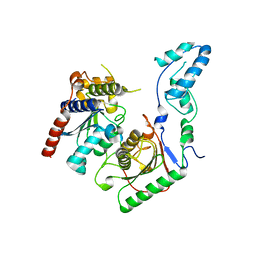 | | Crystal structure of the yeast heterodimeric ADAT2/3 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase subunit TAD2, tRNA-specific adenosine deaminase subunit TAD3 | | Authors: | Xie, W, Liu, X, Chen, R, Sun, Y, Chen, R, Zhou, J, Tian, Q. | | Deposit date: | 2020-04-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the yeast heterodimeric ADAT2/3 deaminase.
Bmc Biol., 18, 2020
|
|
8KI6
 
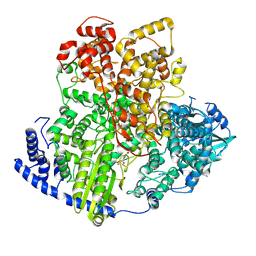 | | Structure of tomato spotted wilt virus L protein binding to Ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
8KI7
 
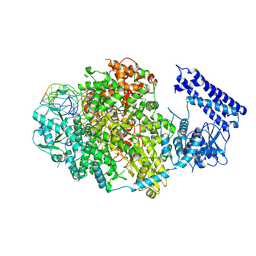 | | Structure of Tomato spotted wilt virus L protein contained endoH domain binding to 3'5'vRNA | | Descriptor: | RNA (5'-R(P*AP*CP*CP*UP*GP*AP*UP*UP*GP*CP*UP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*GP*CP*AP*AP*UP*CP*A)-3'), RNA (5'-R(P*GP*CP*AP*AP*UP*CP*AP*GP*G)-3'), ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
8KIA
 
 | | Tomato spotted wilt virus L protein (apo state) | | Descriptor: | RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
8KI8
 
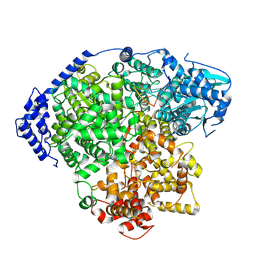 | | structure of Tomato spotted wilt virus L protein binding to 5'vRNA | | Descriptor: | RNA (5'-R(P*AP*GP*AP*GP*CP*AP*AP*UP*CP*A)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
8KI9
 
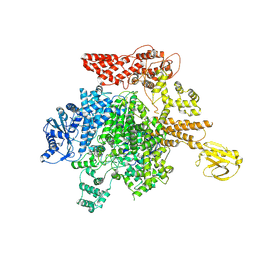 | |
7E1L
 
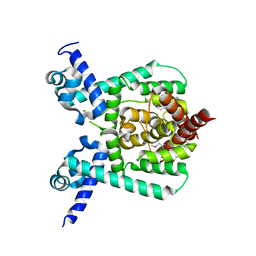 | | Crystal structure of apo form PhlH | | Descriptor: | DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7E1N
 
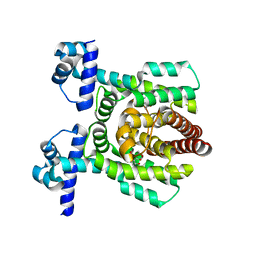 | | Crystal structure of PhlH in complex with 2,4-diacetylphloroglucinol | | Descriptor: | 2,4-bis[(1R)-1-oxidanylethyl]benzene-1,3,5-triol, DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7XUK
 
 | |
7XUN
 
 | |
7XUO
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with cisplatin in presence of ATOX1 | | Descriptor: | Copper-transporting ATPase 2, PLATINUM (II) ION | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7XUM
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with Cu+ in presence of ATOX1 | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 2 | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7XXB
 
 | | IAA bound state of AtPIN3 | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-05-29 | | Release date: | 2022-08-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
8IRL
 
 | | Apo state of Arabidopsis AZG1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRN
 
 | | 6-BAP bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-BENZYL-9H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRP
 
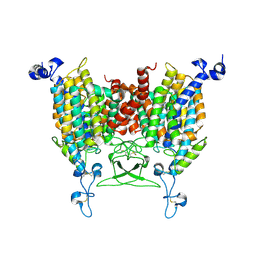 | | kinetin bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRM
 
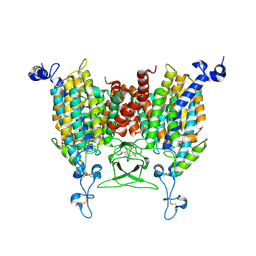 | | Endogenous substrate adenine bound state of Arabidopsis AZG1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
