1PM3
 
 | |
6XVW
 
 | |
6EQS
 
 | | Human Sirt5 in complex with stalled peptidylimidate intermediate of inhibitory compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-BUTANEDIOL, 3-[[(~{Z})-~{C}-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]sulfanyl-~{N}-[(5~{S})-6-[[(2~{S})-3-(1~{H}-indol-3-yl)-1-oxidanylidene-1-(propan-2-ylamino)propan-2-yl]amino]-6-oxidanylidene-5-(phenylmethoxycarbonylamino)hexyl]carbonimidoyl]amino]propanoic acid, ... | | Authors: | Pannek, M, Steegborn, C. | | Deposit date: | 2017-10-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Mechanism-Based Inhibitors of the Human Sirtuin 5 Deacylase: Structure-Activity Relationship, Biostructural, and Kinetic Insight.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4UST
 
 | |
6XV1
 
 | | Human Sirt6 13-308 in complex with ADP-ribose and the activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, NAD-dependent protein deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2020-01-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding site for activator MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
6YII
 
 | |
6YP4
 
 | |
6XVG
 
 | | Human Sirt6 3-318 in complex with ADP-ribose and the activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, GLYCEROL, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2020-01-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding site for activator MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
6XV6
 
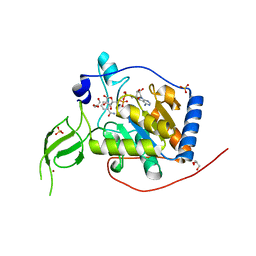 | | Human Sirt6 3-318 in complex with ADP-ribose | | Descriptor: | GLYCEROL, NAD-dependent protein deacetylase sirtuin-6, NICOTINAMIDE, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2020-01-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Binding site for activator MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
6XUY
 
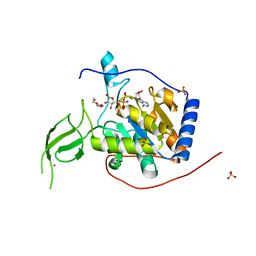 | | Human Sirt6 13-308 in complex with ADP-ribose | | Descriptor: | NAD-dependent protein deacetylase sirtuin-6, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2020-01-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Binding site for activator MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
5MAT
 
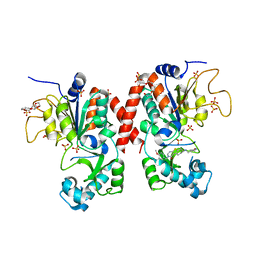 | | Structure of human Sirtuin 2 in complex with a selective thienopyrimidinone based inhibitor | | Descriptor: | (7~{R})-7-[(3,5-dimethyl-1,2-oxazol-4-yl)methylamino]-3-[(4-methoxynaphthalen-1-yl)methyl]-5,6,7,8-tetrahydro-[1]benzothiolo[2,3-d]pyrimidin-4-one, HEXAETHYLENE GLYCOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Moniot, S, Steegborn, C. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Thienopyrimidinone Based Sirtuin-2 (SIRT2)-Selective Inhibitors Bind in the Ligand Induced Selectivity Pocket.
J. Med. Chem., 60, 2017
|
|
5MAR
 
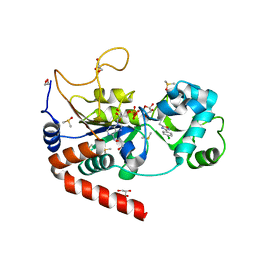 | | Structure of human SIRT2 in complex with 1,2,4-Oxadiazole inhibitor and ADP ribose. | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(4-chlorophenyl)-1,2,4-oxadiazol-5-yl]propan-1-ol, ACETATE ION, ... | | Authors: | Moniot, S, Steegborn, C. | | Deposit date: | 2016-11-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Development of 1,2,4-Oxadiazoles as Potent and Selective Inhibitors of the Human Deacetylase Sirtuin 2: Structure-Activity Relationship, X-ray Crystal Structure, and Anticancer Activity.
J. Med. Chem., 60, 2017
|
|
5OJN
 
 | |
4CLF
 
 | | Crystal structure of human soluble Adenylyl Cyclase (Apo form) | | Descriptor: | ACETATE ION, ADENYLATE CYCLASE TYPE 10, GLYCEROL | | Authors: | Kleinboelting, S, Moniot, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-03-05 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8AK5
 
 | |
8AKB
 
 | |
8AKF
 
 | |
8ANC
 
 | |
8AKD
 
 | |
8AKE
 
 | |
8ANB
 
 | |
8AK8
 
 | |
8AKG
 
 | |
8AK7
 
 | |
8AK6
 
 | |
