1M0B
 
 | | HIV-1 protease in complex with an ethyleneamine inhibitor | | Descriptor: | GLYCEROL, N-{(3S)-3-[(tert-butoxycarbonyl)amino]-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Petrokova, H, Hasek, J, Dohnalek, J. | | Deposit date: | 2002-06-12 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of hydroxyl group and R/S configuration of isostere in binding properties of HIV-1 protease inhibitors
Eur.J.Biochem., 271, 2004
|
|
8QJP
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with uridine - 5'- monophosphate | | Descriptor: | GLYCEROL, PHOSPHATE ION, S1/P1 Nuclease, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJM
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, PENTAETHYLENE GLYCOL, S1/P1 Nuclease, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJL
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia | | Descriptor: | GLYCEROL, S1/P1 Nuclease, SULFATE ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJO
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with guanosine-5'-monophosphate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJN
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with adenosine-5'-monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
8QJQ
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with cytidine - 5' - monophosphate as an inhibitor. | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
9EMA
 
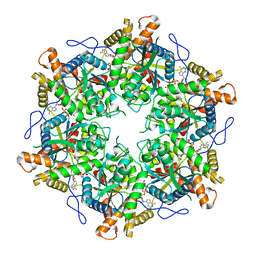
 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
9EMC
 
 | |
1LZQ
 
 | | Crystal structure of the complex of mutant HIV-1 protease (A71V, V82T, I84V) with an ethylenamine peptidomimetic inhibitor BOC-PHE-PSI[CH2CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | BETA-MERCAPTOETHANOL, N-{(3S)-3-[(tert-butoxycarbonyl)amino]-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Hasek, J, Dohnalek, J, Petrokova, H, Buchtelova, E, Soucek, M, Majer, P, Uhlikova, T, Konvalinka, J. | | Deposit date: | 2002-06-11 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Ethylenamine Inhibitor Binds Tightly to Both Wild Type and Mutant HIV-1 Proteases. Structure and Energy Study
J.Med.Chem., 46, 2003
|
|
1YQ2
 
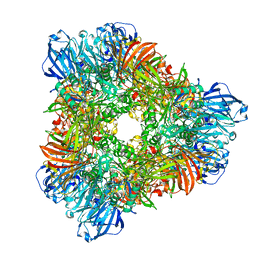 | | beta-galactosidase from Arthrobacter sp. C2-2 (isoenzyme C2-2-1) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Skalova, T, Dohnalek, J, Spiwok, V, Lipovova, P, Vondrackova, E, Petrokova, H, Strnad, H, Kralova, B, Hasek, J. | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cold-active beta-Galactosidase from Arthrobacter sp. C2-2 Forms Compact 660kDa Hexamers: Crystal Structure at 1.9A Resolution
J.Mol.Biol., 353, 2005
|
|
1RCY
 
 | | RUSTICYANIN (RC) FROM THIOBACILLUS FERROOXIDANS | | Descriptor: | COPPER (II) ION, RUSTICYANIN | | Authors: | Walter, R.L, Friedman, A.M, Ealick, S.E, Blake II, R.C, Proctor, P, Shoham, M. | | Deposit date: | 1996-04-10 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability.
J.Mol.Biol., 263, 1996
|
|
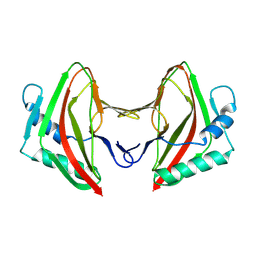
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8AG3
 
 | |
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
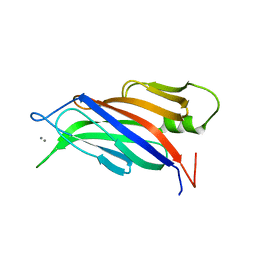
5A51
 
 | |
5A52
 
 | |
5A4X
 
 | |
5A50
 
 | |
4JDG
 
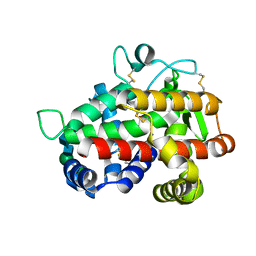 | | Structure of Tomato Bifunctional Nuclease TBN1, variant N211D | | Descriptor: | Nuclease, PHOSPHATE ION, ZINC ION, ... | | Authors: | Stransky, J, Dohnalek, J, Koval, T, Podzimek, T, Lipovova, P, Matousek, J. | | Deposit date: | 2013-02-25 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Phosphate binding in the active centre of tomato multifunctional nuclease TBN1 and analysis of superhelix formation by the enzyme
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5NNL
 
 | |
7AHO
 
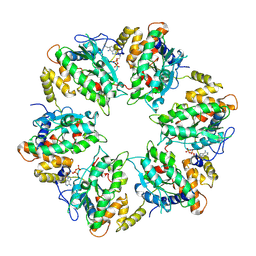 | | RUVBL1-RUVBL2 heterohexameric ring after binding of RNA helicase DHX34 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Lopez-Perrote, A, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2020-09-25 | | Release date: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Regulation of RUVBL1-RUVBL2 AAA-ATPases by the nonsense-mediated mRNA decay factor DHX34, as evidenced by Cryo-EM.
Elife, 9, 2020
|
|
