7KNB
 
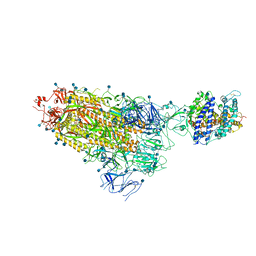 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMS
 
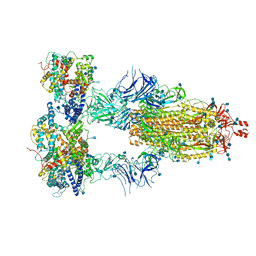 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMZ
 
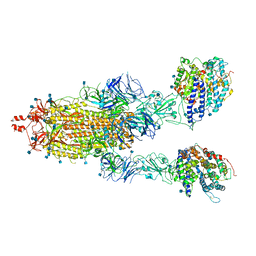 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
1BVA
 
 | | MANGANESE BINDING MUTANT IN CYTOCHROME C PEROXIDASE | | Descriptor: | MANGANESE (II) ION, PROTEIN (CYTOCHROME C PEROXIDASE), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wilcox, S.K, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Rational design of a functional metalloenzyme: introduction of a site for manganese binding and oxidation into a heme peroxidase.
Biochemistry, 37, 1998
|
|
6MTO
 
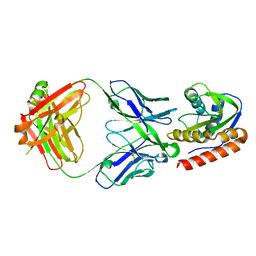 | | Crystal structure of VRC42.01 Fab in complex with T117-F MPER scaffold | | Descriptor: | Antibody VRC42.01 Fab heavy chain, Antibody VRC42.01 Fab light chain, VRC42 epitope T117-F scaffold | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MTR
 
 | | Crystal structure of VRC43.01 Fab | | Descriptor: | Antibody VRC43.01 Fab heavy chain, Antibody VRC43.01 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MTT
 
 | | Crystal structure of VRC46.01 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC46.01 Fab heavy chain, Antibody VRC46.01 Fb light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
5K6F
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-19 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0 | | Authors: | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6MTQ
 
 | | Crystal structure of VRC42.N1 Fab in complex with T117-F MPER scaffold | | Descriptor: | Antibody VRC42.N1 Fab heavy chain, Antibody VRC42.N1 Fab light chain, VRC42 epitope T117-F scaffold | | Authors: | Kwon, Y.D, Law, W.H, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
5K6B
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9 DS-Cav1 variant. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, SULFATE ION | | Authors: | Joyce, M.G, Zhang, B, Rundlet, E.J, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6MTS
 
 | | Crystal structure of VRC43.03 Fab | | Descriptor: | Antibody VRC43.03 Fab heavy chain, Antibody VRC43.03 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MTP
 
 | | Crystal structure of VRC42.04 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC42.04 Fab heavy chain, Antibody VRC42.04 Fab light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
5K6G
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-24 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | Authors: | Joyce, M.G, Zhang, B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5K6C
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0, SULFATE ION | | Authors: | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.576 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5K6H
 
 | |
6MQC
 
 | |
6MPH
 
 | | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DF1W-a.01 Light chain, ... | | Authors: | Acharya, P, Xu, K, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6N16
 
 | |
6N1W
 
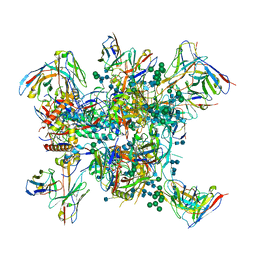 | |
6N1V
 
 | |
6MQR
 
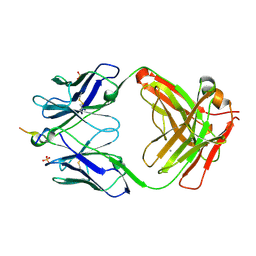 | | Vaccine-elicited NHP FP-targeting neutralizing antibody 0PV-a.01 in complex with FP (residue 512-519) | | Descriptor: | CALCIUM ION, HIV fusion peptide residue 512-519, SULFATE ION, ... | | Authors: | Xu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6MQE
 
 | |
6MQS
 
 | |
6MQM
 
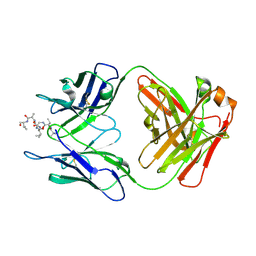 | |
