6HVM
 
 | | Structural characterization of CdaA-APO | | Descriptor: | CHLORIDE ION, Diadenylate cyclase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
3V0X
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bovine trypsin variants X(tripleGlu217Phe227) in complexes with small molecule inhibitor
To be Published
|
|
6HVN
 
 | | CdaA-APO Y187A Mutant | | Descriptor: | CHLORIDE ION, Diadenylate cyclase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
6HVL
 
 | | CdaA complex with c-di-AMP and AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE MONOPHOSPHATE, COBALT (II) ION, ... | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
1KGI
 
 | | Rat transthyretin (also called prealbumin) complex with 3,3',5,5'-tetraiodothyroacetic acid (t4ac) | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, TRANSTHYRETIN | | Authors: | Wojtczak, A, Neumann, P, Muziol, T, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 2001-11-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex of rat transthyretin with tetraiodothyroacetic acid refined at 2.1 and 1.8 A resolution.
Acta Biochim.Pol., 48, 2001
|
|
1KGJ
 
 | | Rat transthyretin (also called prealbumin) complex with 3',5'-dibromoflavone (EMD21388) | | Descriptor: | 6,4'-DIHYDROXY-3-METHYL-3',5'-DIBROMOFLAVONE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Neumann, P, Muziol, T, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 2001-11-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of binding interactions of dibromoflavonoids with transthyretin.
Acta Biochim.Pol., 48, 2001
|
|
6F2C
 
 | | Methylglyoxal synthase MgsA from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, Methylglyoxal synthase | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-11-24 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the regulatory interaction of the methylglyoxal synthase MgsA with the carbon flux regulator Crh inBacillus subtilis.
J. Biol. Chem., 293, 2018
|
|
6FAC
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FA5
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hamann, F, Schmitt, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAA
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHANOL, GLYCEROL, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6H42
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
6H45
 
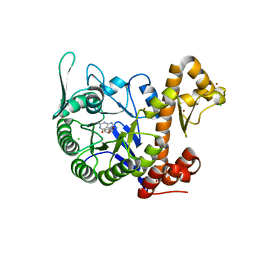 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
5O5O
 
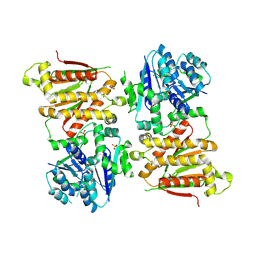 | | X-ray crystal structure of RapZ from Escherichia coli (P32 space group) | | Descriptor: | RNase adapter protein RapZ, SULFATE ION | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
3E07
 
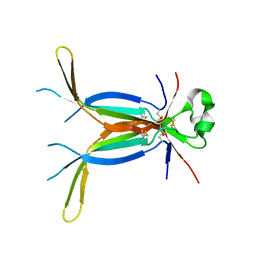 | | Crystal structure of spatzle cystine knot | | Descriptor: | GLYCEROL, Protein spaetzle | | Authors: | Hoffmann, A, Funkner, A, Neumann, P, Juhnke, S, Walther, M, Schierhorn, A, Weininger, U, Balbach, J, Reuter, G, Stubbs, M.T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biophysical Characterization of Refolded Drosophila Spatzle, a Cystine Knot Protein, Reveals Distinct Properties of Three Isoforms
J.Biol.Chem., 283, 2008
|
|
3E7W
 
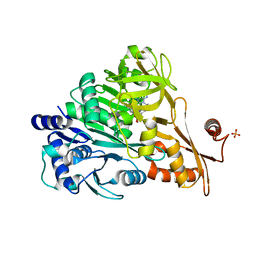 | | Crystal structure of DLTA: Implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1, PHOSPHATE ION | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
3E7X
 
 | | Crystal structure of DLTA: implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1 | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
3EY6
 
 | | Crystal structure of the FK506-binding domain of human FKBP38 | | Descriptor: | FK506-binding protein 8 | | Authors: | Parthier, C, Maestre-Martinez, M, Neumann, P, Edlich, F, Fischer, G, Luecke, C, Stubbs, M.T. | | Deposit date: | 2008-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A charge-sensitive loop in the FKBP38 catalytic domain modulates Bcl-2 binding.
J.Mol.Recognit., 24, 2011
|
|
5O5S
 
 | | X-ray crystal structure of the RapZ C-terminal domain from Escherichia coli | | Descriptor: | MALONATE ION, RNase adapter protein RapZ | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
4NHO
 
 | | Structure of the spliceosomal DEAD-box protein Prp28 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Moehlmann, S, Neumann, P, Ficner, R. | | Deposit date: | 2013-11-05 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5O5Q
 
 | | X-ray crystal structure of RapZ from Escherichia coli (P3221 space group) | | Descriptor: | RNase adapter protein RapZ, SULFATE ION | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
5LZC
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the codon reading state (CR) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZE
 
 | | Structure of the 70S ribosome with Sec-tRNASec in the classical pre-translocation state (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZB
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZD
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the GTPase activated state (GA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5MED
 
 | | Cyanothece lipoxygenase 2 (CspLOX2) | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, 1-BUTANOL, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
