6M1W
 
 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | Amidohydrolase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
6M2I
 
 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
6KZ7
 
 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
7W8Y
 
 | | DMSPP- and Naplha-Me-Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-dimethylallyltryptophan synthase, ... | | Authors: | Suemune, H, Nagano, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8V
 
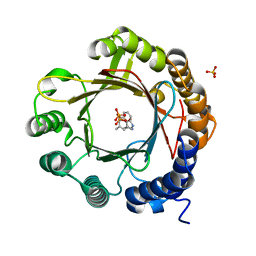 | | DMSPP- and Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, SULFATE ION, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8X
 
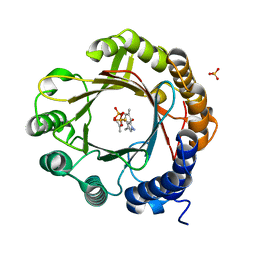 | | DMSPP- and 6-Me-Trp-bound dimethylallyl tryptophan synthase, IptA | | Descriptor: | (2S)-2-azanyl-3-(6-methyl-1H-indol-3-yl)propanoic acid, 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8W
 
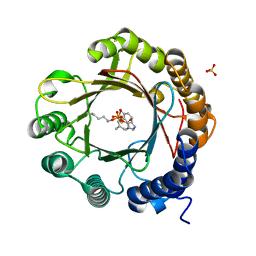 | | DMSPP- and 5-Me-Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 5-methyl-L-tryptophan, 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8U
 
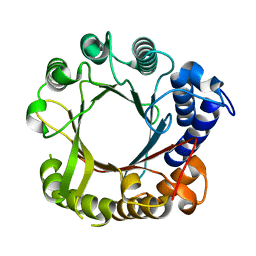 | | Crystal Structure of Indole Prenyltransferase IptA | | Descriptor: | 6-dimethylallyltryptophan synthase | | Authors: | Suemune, H, Nagano, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
3A56
 
 | |
3A54
 
 | |
2ZK9
 
 | | Crystal Structure of Protein-glutaminase | | Descriptor: | GLYCEROL, Protein-glutaminase, SODIUM ION | | Authors: | Hashizume, R. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-17 | | Last modified: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of protein glutaminase and its pro forms converted into enzyme-substrate complex
J.Biol.Chem., 286, 2011
|
|
3A55
 
 | |
3AJ3
 
 | | Crystal structure of selenomethionine substituted 4-pyridoxolactonase from Mesorhizobium loti | | Descriptor: | 4-pyridoxolactonase, PHOSPHATE ION, ZINC ION | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mikami, B, Yagi, T. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
