5FL2
 
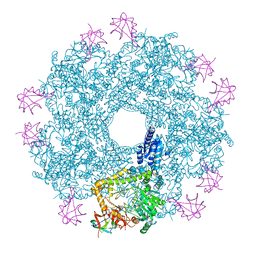 | | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
7YTF
 
 | | Structure of OCPx2 from Nostoc flagelliforme CCNUN1 | | Descriptor: | Ketosteroid isomerase-related protein, beta,beta-carotene-4,4'-dione | | Authors: | Yang, Y.W, Chen, S.Z, Liu, K, Chen, M, Qiu, B.S. | | Deposit date: | 2022-08-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional specialization of expanded orange carotenoid protein paralogs in subaerial Nostoc species.
Plant Physiol., 192, 2023
|
|
7YTH
 
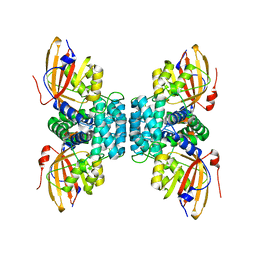 | | Structure of OCPx1 from Nostoc flagelliforme CCNUN1 | | Descriptor: | Ketosteroid isomerase-related protein | | Authors: | Yang, Y.W, Liu, K, Chen, S.Z, Chen, M, Qiu, B.S. | | Deposit date: | 2022-08-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Functional specialization of expanded orange carotenoid protein paralogs in subaerial Nostoc species.
Plant Physiol., 192, 2023
|
|
4R3H
 
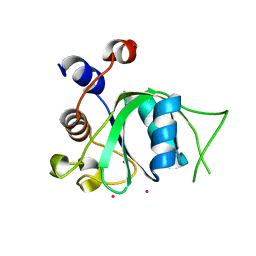 | | The crystal structure of an apo RNA binding protein | | Descriptor: | SULFATE ION, UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Xu, C, Liu, K, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
4RCI
 
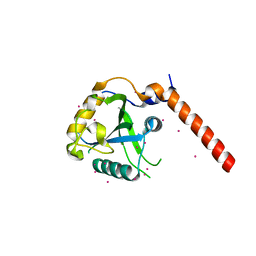 | | Crystal structure of YTHDF1 YTH domain | | Descriptor: | UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Xu, C, Tempel, W, Liu, K, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
4R3I
 
 | | The crystal structure of an RNA complex | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Tempel, W, Xu, C, Liu, K, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
4UPB
 
 | | Electron cryo-microscopy of the complex formed between the hexameric ATPase RavA and the decameric inducible decarboxylase LdcI | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
4UPF
 
 | | Assembly principles of the unique cage formed by the ATPase RavA hexamer and the lysine decarboxylase LdcI decamer | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
1R2L
 
 | | A parallel stranded DNA duplex with an A-G mismatch base-pair | | Descriptor: | 5'-D(P*CP*CP*TP*AP*TP*GP*AP*AP*AP*TP*CP*C)-3', DNA (5'-D(P*(DNR)P*(DNR)P*DAP*DTP*DAP*DAP*DTP*DTP*DTP*DAP*(DNR)P*(DNR))-3') | | Authors: | Venkitakrishnan, R.P, Bhaumik, S.R, Chary, K.V.R, govil, G, Liu, K, Howard, F.B, Miles, T.H. | | Deposit date: | 2003-09-29 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A parallel stranded DNA duplex with an A-G mismatch base-pair: (CCATAATTTACC:CCTATGAAATCC)
RECENT TRENDS IN BIOPHYS.RES., 2004
|
|
5I8E
 
 | | Crystal Structure of Broadly Neutralizing HIV-1 Fusion Peptide-Targeting Antibody VRC34.01 Fab | | Descriptor: | VRC34.01 Fab heavy chain, VRC34.01 Fab light chain, ZINC ION | | Authors: | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
2ZGJ
 
 | | Crystal Structure of D86N-GzmM Complexed with Its Optimal Synthesized Substrate | | Descriptor: | Granzyme M, SSGKVPLS, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZGH
 
 | | Crystal Structure of active granzyme M bound to its product | | Descriptor: | Granzyme M, SSGKVPL, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
5I8C
 
 | | Crystal Structure of HIV-1 Clade A BG505 Fusion Peptide (residue 512-520) in Complex with Broadly Neutralizing Antibody VRC34.01 Fab | | Descriptor: | HIV-1 Clade A BG505 Fusion Peptide (residue 512-520), VRC34.01 Fab heavy chain, VRC34.01 Fab light chain | | Authors: | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
2ZGC
 
 | | Crystal Structure of Active Human Granzyme M | | Descriptor: | Granzyme M, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZKS
 
 | | Structural insights into the proteolytic machinery of apoptosis-inducing Granzyme M | | Descriptor: | Granzyme M, SULFATE ION, hGzmM inhibitor | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Yang, X, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-03-28 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
8K8A
 
 | | Crystal structure of NFIL3 in complex with TTACGTAA DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*G)-3'), Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K89
 
 | | Crystal structure of NFIL3 | | Descriptor: | Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K8D
 
 | |
8K86
 
 | | Crystal structure of NFIL3 in complex with TTATGTAA DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*TP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*AP*CP*AP*TP*AP*AP*TP*G)-3'), Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K8C
 
 | | Crystal structure of C/EBPalpha BZIP domain bound to a high affinity DNA | | Descriptor: | CCAAT/enhancer-binding protein alpha, DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*GP*A)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*GP*T)-3'), ... | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8WG3
 
 | | mouse TMEM63b in LMNG-CHS micelle | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
8WG4
 
 | | mouse TMEM63b in DDM-CHS micelle with YN9303-24 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
5XK4
 
 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XK5
 
 | | Relaxed state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Xu, D, Zhou, G, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7TT9
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y34F variant | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, J.E, Liu, K, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y34F variant
To Be Published
|
|
