1LUG
 
 | | Full Matrix Error Analysis of Carbonic Anhydrase | | Descriptor: | (4-SULFAMOYL-PHENYL)-THIOCARBAMIC ACID O-(2-THIOPHEN-3-YL-ETHYL) ESTER, Carbonic Anhydrase II, GLYCEROL, ... | | Authors: | Merritt, E.A, Le Trong, I, Behnke, C.A. | | Deposit date: | 2002-05-22 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution studies of carbonic anhydrase II.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1DF8
 
 | | S45A MUTANT OF STREPTAVIDIN IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Hyre, D.E, Le Trong, I, Freitag, S, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 1999-11-18 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Ser45 plays an important role in managing both the equilibrium and transition state energetics of the streptavidin-biotin system.
Protein Sci., 9, 2000
|
|
1EV4
 
 | | RAT GLUTATHIONE S-TRANSFERASE A1-1: MUTANT W21F/F220Y WITH GSO3 BOUND | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1-1, GLUTATHIONE SULFONIC ACID, SULFATE ION | | Authors: | Adman, E.T, Le Trong, I, Stenkamp, R.E, Nieslanik, B.S, Dietze, E.C, Tai, G, Ibarra, C, Atkins, W.M. | | Deposit date: | 2000-04-19 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Localization of the C-terminus of rat glutathione S-transferase A1-1: crystal structure of mutants W21F and W21F/F220Y.
Proteins, 42, 2001
|
|
1EV9
 
 | | RAT GLUTATHIONE S-TRANSFERASE A1-1 MUTANT W21F WITH GSO3 BOUND | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1-1, GLUTATHIONE SULFONIC ACID, SULFATE ION | | Authors: | Adman, E.T, Le Trong, I, Stenkamp, R.E, Nieslanik, B.S, Dietze, E.C, Tai, G, Ibarra, C, Atkins, W.M. | | Deposit date: | 2000-04-19 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Localization of the C-terminus of rat glutathione S-transferase A1-1: crystal structure of mutants W21F and W21F/F220Y.
Proteins, 42, 2001
|
|
1DVB
 
 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, RUBRERYTHRIN, ZINC ION | | Authors: | Sieker, L.C, Holmes, M, Le Trong, I, Turley, S, Liu, M.Y, Legall, J, Stenkamp, R.E. | | Deposit date: | 2000-01-20 | | Release date: | 2000-11-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A crystal structure of the "as isolated" rubrerythrin from Desulfovibrio vulgaris: some surprising results.
J.Biol.Inorg.Chem., 5, 2000
|
|
1SWT
 
 | | CORE-STREPTAVIDIN MUTANT D128A IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Freitag, S, Le Trong, I, Chu, V, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-22 | | Release date: | 1999-07-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural snapshot of an intermediate on the streptavidin-biotin dissociation pathway.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1SWS
 
 | | CORE-STREPTAVIDIN MUTANT D128A AT PH 4.5 | | Descriptor: | PROTEIN (STREPTAVIDIN) | | Authors: | Freitag, S, Chu, V, Le Trong, I, Klumb, L.A, To, R, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-22 | | Release date: | 1999-07-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural snapshot of an intermediate on the streptavidin-biotin dissociation pathway.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1SWU
 
 | | STREPTAVIDIN MUTANT Y43F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-12 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Atomic resolution structure of biotin-free Tyr43Phe streptavidin: what is in the binding site?
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1SWE
 
 | | APO-CORE-STREPTAVIDIN IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWD
 
 | | APO-CORE-STREPTAVIDIN IN COMPLEX WITH BIOTIN (TWO UNOCCUPIED BINDING SITES) AT PH 4.5 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWC
 
 | | APO-CORE-STREPTAVIDIN AT PH 4.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWB
 
 | | APO-CORE-STREPTAVIDIN AT PH 7.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWA
 
 | | APO-CORE-STREPTAVIDIN AT PH 4.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1TH1
 
 | | Beta-catenin in complex with a phosphorylated APC 20aa repeat fragment | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin | | Authors: | Xing, Y, Clements, W.K, Le Trong, I, Hinds, T.R, Stenkamp, R, Kimelman, D, Xu, W. | | Deposit date: | 2004-05-31 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a beta-Catenin/APC Complex Reveals a Critical Role for APC Phosphorylation in APC Function.
Mol.Cell, 15, 2004
|
|
2QCB
 
 | | T7-tagged full-length streptavidin complexed with ruthenium ligand | | Descriptor: | N-(4-{[(2-AMINOETHYL)AMINO]SULFONYL}PHENYL)-5-[(3AS,4S,6AR)-2-OXOHEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL]PENTANAMIDE-(1,2,3,4,5,6-ETA)-BENZENE-CHLORO-RUTHENIUM(III), Streptavidin | | Authors: | Le Trong, I, Creus, M, Pordea, A, Ward, T.R, Stenkamp, R.E. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and designed evolution of an artificial transfer hydrogenase
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
4BVU
 
 | | Structure of Shigella effector OspG in complex with host UbcH5c- Ubiquitin conjugate | | Descriptor: | PROTEIN KINASE OSPG, UBIQUITIN, UBIQUITIN-CONJUGATING ENZYME E2 D3 | | Authors: | Pruneda, J.N, LeTrong, I, Stenkamp, R.E, Klevit, R.E, Brzovic, P.S. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | E2~Ub Conjugates Regulate the Kinase Activity of Shigella Effector Ospg During Pathogenesis.
Embo J., 33, 2014
|
|
1BEA
 
 | | BIFUNCTIONAL HAGEMAN FACTOR/AMYLASE INHIBITOR FROM MAIZE | | Descriptor: | BIFUNCTIONAL AMYLASE/SERINE PROTEASE INHIBITOR | | Authors: | Behnke, C.A, Yee, V.C, Le Trong, I, Pedersen, L.C, Stenkamp, R.E, Kim, S.S, Reeck, G.R, Teller, D.C. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution.
Biochemistry, 37, 1998
|
|
1BFA
 
 | | RECOMBINANT BIFUNCTIONAL HAGEMAN FACTOR/AMYLASE INHIBITOR FROM MAIZE | | Descriptor: | BIFUNCTIONAL AMYLASE/SERINE PROTEASE INHIBITOR | | Authors: | Behnke, C.A, Yee, V.C, Le Trong, I, Pedersen, L.C, Stenkamp, R.E, Kim, S.S, Reeck, G.R, Teller, D.C. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution.
Biochemistry, 37, 1998
|
|
6VMT
 
 | |
6VYE
 
 | |
1GGT
 
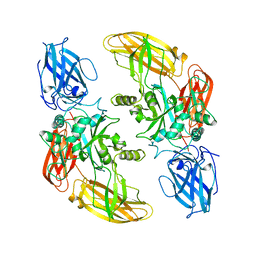 | | THREE-DIMENSIONAL STRUCTURE OF A TRANSGLUTAMINASE: HUMAN BLOOD COAGULATION FACTOR XIII | | Descriptor: | COAGULATION FACTOR XIII | | Authors: | Yee, V.C, Pedersen, L.C, Trong, I.L, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1994-01-25 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Three-dimensional structure of a transglutaminase: human blood coagulation factor XIII.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
2AR1
 
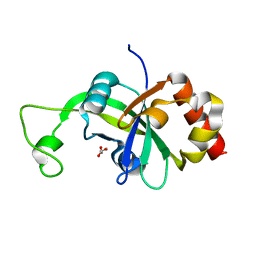 | |
2I35
 
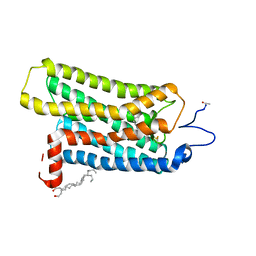 | | Crystal structure of rhombohedral crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I36
 
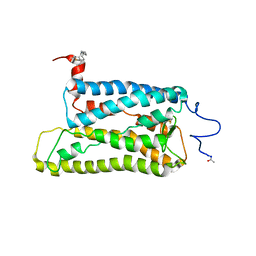 | | Crystal structure of trigonal crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, Rhodopsin, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I37
 
 | | Crystal structure of a photoactivated rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Rhodopsin, ... | | Authors: | Lodowski, D.T, Stenkamp, R.E, Salom, D, Le Trong, I, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
