1CEX
 
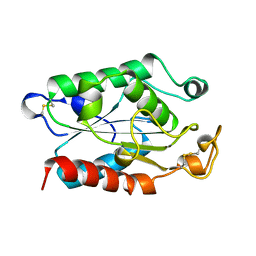 | | STRUCTURE OF CUTINASE | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Czjzek, M, Lamzin, V, Nicolas, A, Cambillau, C. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: stereochemical analysis.
J.Mol.Biol., 268, 1997
|
|
1QWD
 
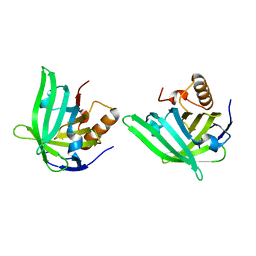 | | CRYSTAL STRUCTURE OF A BACTERIAL LIPOCALIN, THE BLC GENE PRODUCT FROM E. COLI | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Campanacci, V, Nurizzo, D, Spinelli, S, Valencia, C, Cambillau, C. | | Deposit date: | 2003-09-02 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding
Febs Lett., 562, 2004
|
|
6EFE
 
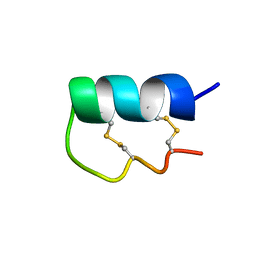 | | NMR Solution Structure of vil14a | | Descriptor: | Kappa-conotoxin vil14a | | Authors: | Dovell, S, Mari, F, Moller, C, Melaun, C. | | Deposit date: | 2018-08-16 | | Release date: | 2018-09-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Definition of the R-superfamily of conotoxins: Structural convergence of helix-loop-helix peptidic scaffolds.
Peptides, 107, 2018
|
|
4V5I
 
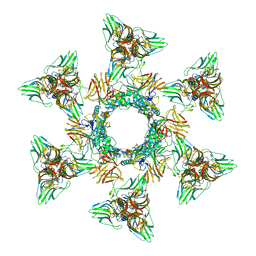 | | Structure of the Phage P2 Baseplate in its Activated Conformation with Ca | | Descriptor: | CALCIUM ION, ORF15, ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.464 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1GT5
 
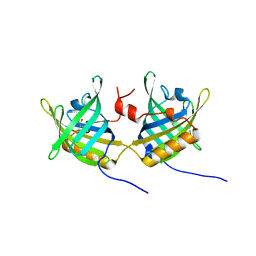 | | Complexe of Bovine Odorant Binding Protein with benzophenone | | Descriptor: | DIPHENYLMETHANONE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1HN2
 
 | | CRYSTAL STRUCTURE OF BOVINE OBP COMPLEXED WITH AMINOANTHRACENE | | Descriptor: | (3R)-oct-1-en-3-ol, ANTHRACEN-1-YLAMINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
3BFA
 
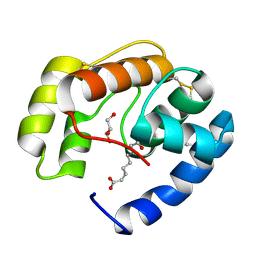 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the Queen mandibular pheromone | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3ZVK
 
 | | Crystal structure of VapBC2 from Rickettsia felis bound to a DNA fragment from their promoter | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTITOXIN OF TOXIN-ANTITOXIN SYSTEM VAPB, DNA, ... | | Authors: | Mate, M.J, Ortiz-Lombardia, M, Cambillau, C. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the DNA-Bound Vapbc2 Antitoxin/Toxin Pair from Rickettsia Felis.
Nucleic Acids Res., 40, 2012
|
|
1XZA
 
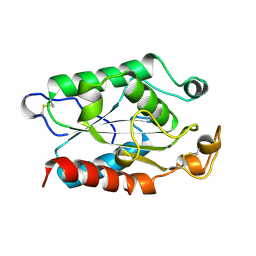 | |
1XZB
 
 | |
1XZE
 
 | |
1QYA
 
 | | CRYSTAL STRUCTURE OF E. COLI PROTEIN YDDE | | Descriptor: | HYPOTHETICAL PROTEIN yddE | | Authors: | Grassick, A, Sulzenbacher, G, Roig-Zamboni, V, Campanacci, V, Cambillau, C, Bourne, Y. | | Deposit date: | 2003-09-10 | | Release date: | 2004-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of E. coli yddE protein reveals a striking homology with diaminopimelate epimerase
Proteins, 55, 2004
|
|
1QY9
 
 | | Crystal structure of E. coli Se-MET protein YDDE | | Descriptor: | GLYCEROL, HYDROXIDE ION, HYPOTHETICAL PROTEIN yddE | | Authors: | Grassick, A, Sulzenbacher, G, Roig-Zamboni, V, Campanacci, V, Cambillau, C, Bourne, Y. | | Deposit date: | 2003-09-10 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of E. coli yddE protein reveals a striking homology with diaminopimelate epimerase
Proteins, 55, 2004
|
|
1XZJ
 
 | |
1XZI
 
 | |
1XZM
 
 | |
1XZG
 
 | |
1XZH
 
 | |
1XZD
 
 | |
1XZF
 
 | |
1H8O
 
 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment. | | Descriptor: | MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
1HLG
 
 | | CRYSTAL STRUCTURE OF HUMAN GASTRIC LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIPASE, ... | | Authors: | Roussel, A, Canaan, S, Verger, R, Cambillau, C. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human gastric lipase and model of lysosomal acid lipase, two lipolytic enzymes of medical interest.
J.Biol.Chem., 274, 1999
|
|
1OXM
 
 | | STRUCTURE OF CUTINASE | | Descriptor: | BUTYL-PHOSPHINIC ACID 2,3-BIS-BUTYLCARBAMOYLOXY-PROPYL ESTER GROUP, CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1996-10-26 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cutinase covalently inhibited by a triglyceride analogue.
Protein Sci., 6, 1997
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
1XZK
 
 | |
