8P60
 
 | | Spraguea lophii ribosome dimer | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2023-05-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (14.3 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
8P5D
 
 | | Spraguea lophii ribosome in the closed conformation by cryo sub tomogram averaging | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2023-05-23 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
8PAJ
 
 | | Crystal Structure of a Squalene-Hopene cyclase from Archangium gephyra | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Worthy, H.L, Isupov, M.N, Littlechild, J.A, Mitchell, D.E. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of a mesophilic Squalene-Hopene Cyclases from Cystobacter fuscus and Archangium gephyra
To Be Published
|
|
8PAK
 
 | | Crystal Structure of a Squalene-Hopene Cyclase from Cystobacter fuscus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Worthy, H.L, Mitchell, D.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of Mesophilic Squalene-Hopene Cyclases from Cystobacter fuscus and Archangium gephyra
To Be Published
|
|
5NFQ
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5NG7
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5O44
 
 | | Crystal structure of unbranched mixed tri-Ubiquitin chain containing K48 and K63 linkages. | | Descriptor: | MAGNESIUM ION, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2017-05-26 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | The Crystal Structure and Conformations of an Unbranched Mixed Tri-Ubiquitin Chain Containing K48 and K63 Linkages.
J. Mol. Biol., 429, 2017
|
|
1T9H
 
 | | The crystal structure of YloQ, a circularly permuted GTPase. | | Descriptor: | ACETATE ION, CALCIUM ION, Probable GTPase engC, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Cladiere, L, Antson, A.A, Isupov, M.N, Seror, S.J, Wilkinson, A.J. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
J.Mol.Biol., 340, 2004
|
|
2WKW
 
 | | Alcaligenes esterase complexed with product analogue | | Descriptor: | CARBOXYLESTERASE, GLYCEROL, SULFATE ION, ... | | Authors: | Bourne, P.C, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Atomic-Resolution Structure of a Novel Bacterial Esterase.
Structure, 8, 2000
|
|
1HL7
 
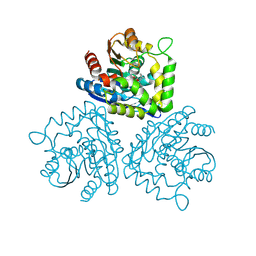 | | Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one | | Descriptor: | 3A,4,7,7A-TETRAHYDRO-BENZO [1,3] DIOXOL-2-ONE, GAMMA LACTAMASE | | Authors: | Line, K, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2003-03-14 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of a (-)Gamma-Lactamase from an Aureobacterium Species Reveals a Tetrahedral Intermediate in the Active Site
J.Mol.Biol., 338, 2004
|
|
2W11
 
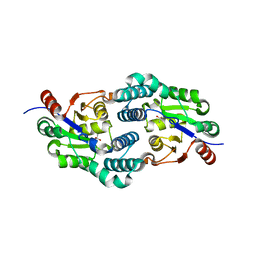 | | Structure of the L-2-haloacid dehalogenase from Sulfolobus tokodaii | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-HALOALKANOIC ACID DEHALOGENASE | | Authors: | Rye, C.A, Isupov, M.N, Lebedev, A.A, Littlechild, J.A. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Studies of a L-Haloacid Dehalogenase from the Thermophilic Archaeon Sulfolobus Tokodaii.
Extremophiles, 13, 2009
|
|
2YN4
 
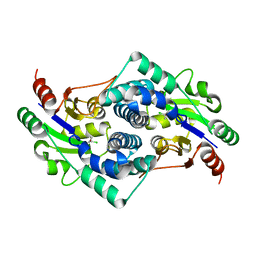 | | L-2-chlorobutryic acid bound complex L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | (2S)-2-chlorobutanoic acid, L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMP
 
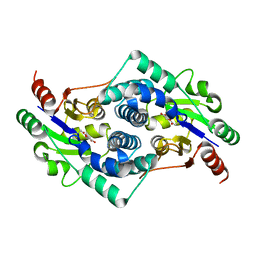 | | Chloroacetic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YML
 
 | | Native L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMQ
 
 | | Chloropropionic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMM
 
 | | Sulfate bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE, SULFATE ION | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
3ZRR
 
 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CALCIUM ION, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AH3
 
 | |
4B9B
 
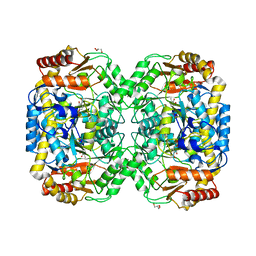 | | The structure of the omega aminotransferase from Pseudomonas aeruginosa | | Descriptor: | BETA-ALANINE-PYRUVATE TRANSAMINASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sayer, C, Isupov, M.N, Westlake, A, Littlechild, J.A. | | Deposit date: | 2012-09-03 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BA4
 
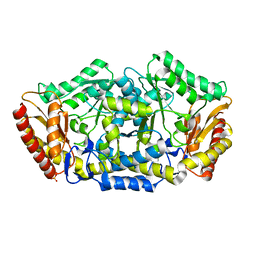 | |
4BA5
 
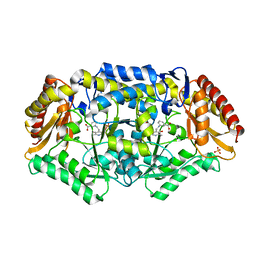 | | Crystal structure of omega-transaminase from Chromobacterium violaceum | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, AMINOTRANSFERASE, SULFATE ION | | Authors: | Sayer, C, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2012-09-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZRP
 
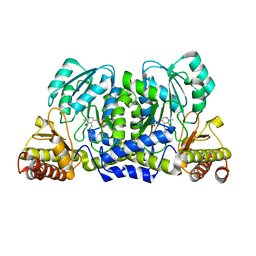 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ZRQ
 
 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4B98
 
 | | The structure of the omega aminotransferase from Pseudomonas aeruginosa | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, BETA-ALANINE--PYRUVATE TRANSAMINASE, CALCIUM ION, ... | | Authors: | Sayer, C, Isupov, M.N, Westlake, A, Littlechild, J.A. | | Deposit date: | 2012-09-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BQN
 
 | | Structural insights into WcbI, a novel polysaccharide biosynthesis enzyme. Native protein. | | Descriptor: | CAPSULAR POLYSACCHARIDE BIOSYNTHESIS PROTEIN, CHLORIDE ION, COENZYME A, ... | | Authors: | Vivoli, M, Ayres, E, Isupov, M.N, Harmer, N.J. | | Deposit date: | 2013-05-31 | | Release date: | 2013-11-06 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Insights Into Wcbi, a Novel Polysaccharide-Biosynthesis Enzyme.
Iucrj, 1, 2014
|
|
