4JFM
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate | | Descriptor: | 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFI
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione | | Descriptor: | 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFJ
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFK
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4LAX
 
 | | Crystal Structure Analysis of FKBP52, Complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
4JFL
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one | | Descriptor: | 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4LAV
 
 | | Crystal Structure Analysis of FKBP52, Crystal Form II | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, SULFATE ION | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
4LAW
 
 | | Crystal Structure Analysis of FKBP52, Crystal Form III | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
4LAY
 
 | | Crystal Structure Analysis of FKBP52, Complex with I63 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
7A6X
 
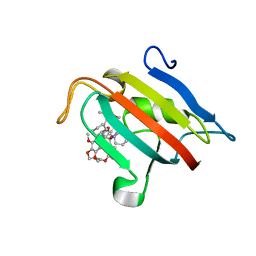 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 56 | | Descriptor: | (2S,9S,12R)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-24,27-dimethoxy-11,18,22-trioxa-4-azatetracyclo[21.2.2.113,17.04,9]octacosa-1(25),13(28),14,16,23,26-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7A6W
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 33-(Z) | | Descriptor: | (2S,9S,12R,20Z)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-28,31-dimethoxy-11,18,23,26-tetraoxa-4-azatetracyclo[25.2.2.113,17.04,9]dotriaconta-1(29),13(32),14,16,20,27,30-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AWX
 
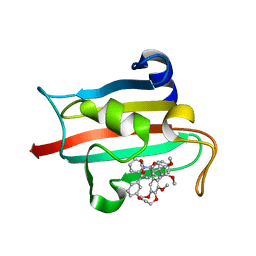 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 55 | | Descriptor: | Macrocyclic SAFit analogue 55, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AWF
 
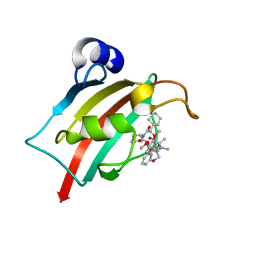 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-11-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7AOU
 
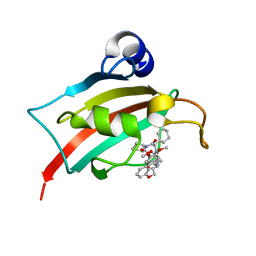 | | The Fk1 domain of FKBP51 in complex with (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone | | Descriptor: | (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7APS
 
 | | The Fk1 domain of FKBP51 in complex with (2S)-2-((1S,5R,6R)-10-((3,5-dichlorophenyl)sulfonyl)-2-oxo-5-vinyl-3,10-diazabicyclo[4.3.1]decan-3-yl)propanoic acid | | Descriptor: | (2~{S})-2-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]propanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
7APT
 
 | | The Fk1 domain of FKBP51 in complex with ((1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-2-oxo-5-vinyl-3,10-diazabicyclo[4.3.1]decan-3-yl)acetic acid | | Descriptor: | 2-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
7APQ
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-(1,3-benzothiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
7B9Y
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 64a | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,24(28),25-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7BA0
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 63 | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,28-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.2.2.113,17.04,9]nonacosa-1(26),13(29),14,16,24,27-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B9Z
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 35-(E) | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,20,24(28),25-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5, isothiocyanate | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8BJE
 
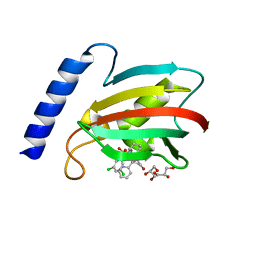 | | A structure of the truncated LpMIP with bound inhibitor JK236. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BJC
 
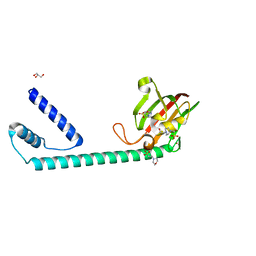 | | Full length structure of the apo-state LpMIP. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-03 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
6SLO
 
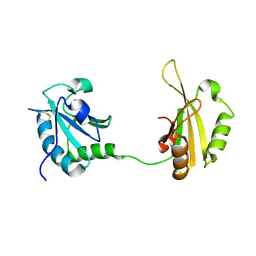 | | Crystal structure of PUF60 UHM domain in complex with 7,8 dimethoxyperphenazine | | Descriptor: | 2-[4-[3-(8-chloranyl-2,3-dimethoxy-phenothiazin-10-yl)propyl]piperazin-1-yl]ethanol, MAGNESIUM ION, Thioredoxin,Poly(U)-binding-splicing factor PUF60 | | Authors: | Jagtap, P.K.A, Kubelka, T, Bach, T, Sattler, M. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of phenothiazine derivatives as UHM-binding inhibitors of early spliceosome assembly.
Nat Commun, 11, 2020
|
|
8BK6
 
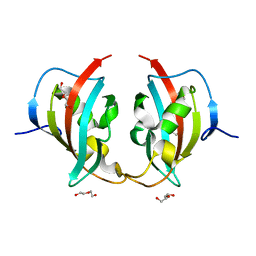 | | A truncated structure of LpMIP with bound inhibitor JK095. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Hellmich, A.U, Goretzki, B. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BJD
 
 | | Full length structure of LpMIP with bound inhibitor JK095 | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
