7OTQ
 
 | | Cryo-EM structure of ALC1/CHD1L bound to a PARylated nucleosome | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (149-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome
Elife, 10, 2021
|
|
5ALI
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 1-[3-[5-(7-aminothiazolo[5,4-d]pyrimidin-2-yl)-1-[[(2S)-tetrahydrofuran-2-yl]methyl]imidazol-4-yl]phenyl]-3-(4-methoxyphenyl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5AB8
 
 | | High resolution X-ray structure of the N-terminal truncated form (residues 1-11) of Mycobacterium tuberculosis HbN | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Pesce, A, Bustamante, J.P, Bidon-Chanal, A, Boechi, L, Estrin, D.A, Luque, F.J, Sebilo, A, Guertin, M, Bolognesi, M, Ascenzi, P, Nardini, M. | | Deposit date: | 2015-08-04 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The N-Terminal Pre-A Region of Mycobacterium Tuberculosis 2/2Hbn Promotes No-Dioxygenase Activity.
FEBS J., 283, 2016
|
|
5ACL
 
 | | Mcg immunoglobulin variable domain with sulfasalazine | | Descriptor: | 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, MCG, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Salwinski, L, Phillips, M.L, Ly, A.T, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain.
Elife, 4, 2015
|
|
5IBX
 
 | | 1.65 Angstrom Crystal Structure of Triosephosphate Isomerase (TIM) from Streptococcus pneumoniae | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.65 Angstrom Crystal Structure of Triosephosphate Isomerase (TIM) from Streptococcus pneumoniae
To Be Published
|
|
5FQN
 
 | |
5A90
 
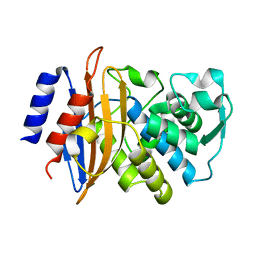 | | 100K Neutron Ligand Free: Exploring the Mechanism of beta-Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | BETA-LACTAMASE CTX-M-97 | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (1.7 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5FWA
 
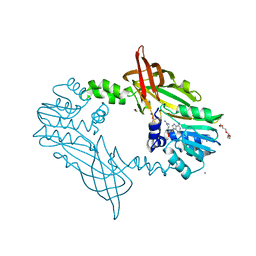 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP1 | | Descriptor: | 1,2-ETHANEDIOL, 9-(6-carbamimidamido-5,6-dideoxy-beta-D-ribo-hexofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5FW5
 
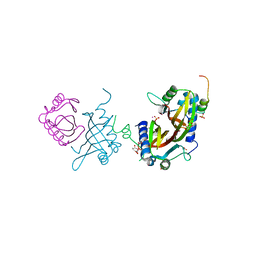 | | Crystal structure of human G3BP1 in complex with Semliki Forest Virus nsP3-25 comprising two FGDF motives | | Descriptor: | ACETATE ION, GLYCEROL, NON-STRUCTURAL PROTEIN 3, ... | | Authors: | Schulte, T, Liu, L, Panas, M.D, Thaa, B, Goette, B, Achour, A, McInerney, G.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication.
Open Biol, 6, 2016
|
|
5FR8
 
 | |
5FM0
 
 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis (PtCl4 derivative) | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PIMELOYL-AMP, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
5A3H
 
 | | 2-DEOXY-2-FLURO-B-D-CELLOBIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
5G0G
 
 | | Crystal structure of Danio rerio HDAC6 CD1 in complex with trichostatin A | | Descriptor: | CHLORIDE ION, HDAC6, SODIUM ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5FRT
 
 | |
5FN0
 
 | | Crystal structure of Pseudomonas fluorescens kynurenine-3- monooxygenase (KMO) in complex with GSK180 | | Descriptor: | 3-(5,6-DICHLORO-2-OXOBENZO[D]OXAZOL-3(2H)-YL)PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, KYNURENINE 3-MONOOXYGENASE | | Authors: | Mole, D.J, Webster, S.P, Uings, I, Zheng, X, Binnie, M, Wilson, K, Hutchinson, J.P, Mirguet, O, Walker, A, Beaufils, B, Ancellin, N, Trottet, L, Beneton, V, Mowat, C.G, Wilkinson, M, Rowland, P, Haslam, C, McBride, A, Homer, N.Z.M, Baily, J.E, Sharp, M.G.F, Garden, O.J, Hughes, J, Howie, S.E.M, Holmes, D, Liddle, J, Iredale, J.P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Kynurenine-3-Monooxygenase Inhibition Prevents Multiple Organ Failure in Rodent Models of Acute Pancreatitis.
Nat.Med. (N.Y.), 22, 2016
|
|
5A59
 
 | | The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A7J
 
 | | Crystal structure of INPP5B in complex with benzene 1,2,4,5- tetrakisphosphate | | Descriptor: | (2,4,5-triphosphonooxyphenyl) dihydrogen phosphate, GLYCEROL, SULFATE ION, ... | | Authors: | Tresaugues, L, Mills, S.J, Silvander, C, Cozier, G, Potter, B.V.L, Nordlund, P. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Type-II Inositol Polyphosphate 5-Phosphatase Inpp5B with Synthetic Inositol Polyphosphate Surrogates Reveal New Mechanistic Insights for the Inositol 5-Phosphatase Family.
Biochemistry, 55, 2016
|
|
5A4W
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5AK3
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, INDAN-2-AMINE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5AIB
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 6-chloro-2H-1,4-benzoxazin-3(4H)-one, BIFUNCTIONAL EPOXIDE HYDROLASE 2, SULFATE ION | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5AL4
 
 | | Crystal structure of TNKS2 in complex with 2-(4-methylpiperazin-1-yl)- 3,4,5,6,7,8-hexahydroquinazolin-4-one | | Descriptor: | 2-(4-METHYLPIPERAZIN-1-YL)-3,4,5,6,7,8-HEXAHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Lehtio, L. | | Deposit date: | 2015-03-06 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Nonplanar Tankyrase Inhibiting Nicotinamide Mimics.
Bioorg.Med.Chem., 23, 2015
|
|
5ALF
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 2-phenyl-N,N-dipropyl-1H-benzimidazole-5-sulfonamide, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5G02
 
 | | Crystal Structure of zebrafish Protein Arginine Methyltransferase 2 with SFG | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LITHIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5ALH
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 4-[[(2-methoxy-5-methyl-phenyl)sulfonylamino]methyl]-4-phenyl-N-(p-tolylmethyl)piperidine-1-carboxamide, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5AKI
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
