1WYO
 
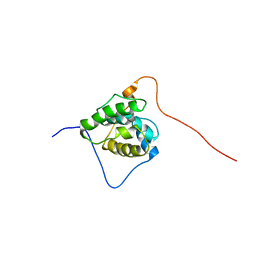 | | Solution structure of the CH domain of human microtubule-associated protein RP/EB family member 3 | | Descriptor: | Microtubule-associated protein RP/EB family member 3 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-15 | | Release date: | 2005-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain of human microtubule-associated protein RP/EB family member 3
To be Published
|
|
1WYR
 
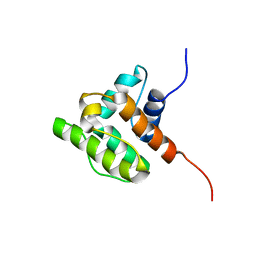 | | Solution structure of the CH domain of human Rho guanine nucleotide exchange factor 6 | | Descriptor: | Rho guanine nucleotide exchange factor 6 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-15 | | Release date: | 2005-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain of human Rho guanine nucleotide exchange factor 6
To be Published
|
|
1WU2
 
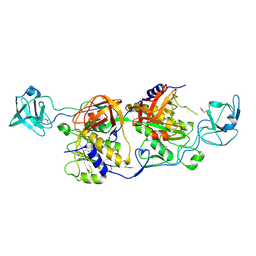 | |
1WYQ
 
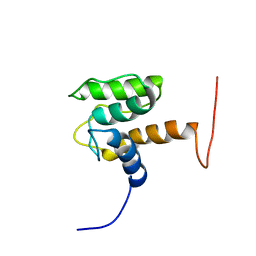 | | Solution structure of the second CH domain of human spectrin beta chain, brain 2 | | Descriptor: | Spectrin beta chain, brain 2 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-15 | | Release date: | 2005-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second CH domain of human spectrin beta chain, brain 2
To be Published
|
|
1X4S
 
 | | Solution structure of zinc finger HIT domain in protein FON | | Descriptor: | ZINC ION, Zinc finger HIT domain containing protein 2 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger HIT domain in protein FON
Protein Sci., 16, 2007
|
|
1X56
 
 | |
1WFK
 
 | |
1WFZ
 
 | | Solution structure of Iron-sulfur cluster protein U (IscU) | | Descriptor: | ZINC ION, nitrogen fixation cluster-like | | Authors: | Nakanishi, T, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Iron-sulfur cluster protein U (IscU)
To be Published
|
|
1WHU
 
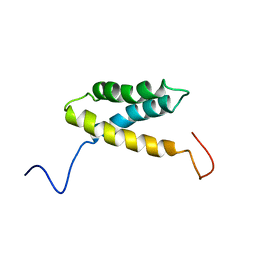 | | Solution structure of the alpha-helical domain from mouse hypothetical PNPase | | Descriptor: | polynucleotide phosphorylase; 3'-5' RNA exonuclease | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the alpha-helical domain from mouse hypothetical PNPase
To be Published
|
|
1WIS
 
 | |
1X62
 
 | |
1WFW
 
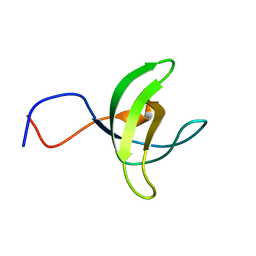 | | Solution structure of SH3 domain of mouse Kalirin-9a protein | | Descriptor: | Kalirin-9a | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3 domain of mouse Kalirin-9a protein
To be Published
|
|
1WIK
 
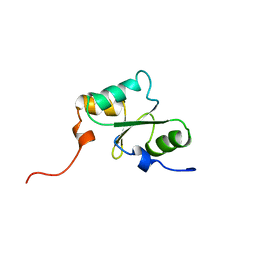 | | Solution Structure of the PICOT homology 2 domain of the mouse PKC-interacting cousin of thioredoxin protein | | Descriptor: | Thioredoxin-like protein 2 | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the PICOT homology 2 domain of the mouse PKC-interacting cousin of thioredoxin protein
To be Published
|
|
1WES
 
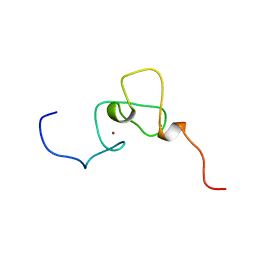 | | Solution structure of PHD domain in inhibitor of growth family, member 1-like | | Descriptor: | ZINC ION, inhibitor of growth family, member 1-like | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in inhibitor of growth family, member 1-like
To be Published
|
|
1WIN
 
 | | Solution Structure of the Band 7 Domain of the mouse Flotillin 2 Protein | | Descriptor: | Flotillin 2 | | Authors: | Miyamoto, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Band 7 Domain of the mouse Flotillin 2 Protein
To be Published
|
|
1WJX
 
 | | Crystal sturucture of TT0801 from Thermus thermophilus | | Descriptor: | POTASSIUM ION, SsrA-binding protein | | Authors: | Bessho, Y, Shibata, R, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1WJZ
 
 | | Soluiotn structure of J-domain of mouse DnaJ like protein | | Descriptor: | 1700030A21Rik protein | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Tochio, N, Tomizawa, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Soluiotn structure of J-domain of mouse DnaJ like protein
To be Published
|
|
1WN2
 
 | |
1WUQ
 
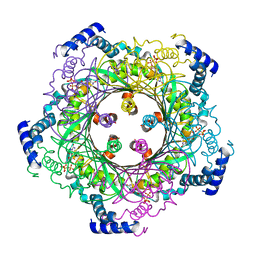 | | Structure of GTP cyclohydrolase I Complexed with 8-oxo-GTP | | Descriptor: | 8-OXO-GUANOSINE-5'-TRIPHOSPHATE, GTP cyclohydrolase I, ZINC ION | | Authors: | Tanaka, Y, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-08 | | Release date: | 2005-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel reaction mechanism of GTP cyclohydrolase I. High-resolution X-ray crystallography of Thermus thermophilus HB8 enzyme complexed with a transition state analogue, the 8-oxoguanine derivative
J.Biochem.(Tokyo), 138, 2005
|
|
1WWQ
 
 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
1WYM
 
 | | Solution structure of the CH domain of human transgelin-2 | | Descriptor: | Transgelin-2 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-15 | | Release date: | 2005-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain of human transgelin-2
To be Published
|
|
1X55
 
 | | Crystal structure of asparaginyl-tRNA synthetase from Pyrococcus horikoshii complexed with asparaginyl-adenylate analogue | | Descriptor: | 5'-O-[N-(L-ASPARAGINYL)SULFAMOYL]ADENOSINE, Asparaginyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Water-assisted Asparagine Recognition by Asparaginyl-tRNA Synthetase.
J.Mol.Biol., 360, 2006
|
|
1WDI
 
 | | Crystal Structure Of TT0907 From Thermus Thermophilus HB8 | | Descriptor: | CITRIC ACID, hypothetical protein TT0907 | | Authors: | Idaka, M, Yagi, H, Murayama, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure Of TT0907 From Thermus Thermophilus HB8
To be published
|
|
1WDT
 
 | | Crystal structure of ttk003000868 from Thermus thermophilus HB8 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, elongation factor G homolog | | Authors: | Wang, H, Takemoto-Hori, C, Murayama, K, Sekine, S, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-17 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ttk003000868 from Thermus thermophilus HB8
To be published
|
|
1WE7
 
 | | Solution structure of Ubiquitin-like domain in SF3a120 | | Descriptor: | Sf3a1 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ubiquitin-like domain in SF3a120
To be Published
|
|
