7K3J
 
 | |
7K3K
 
 | |
7K3L
 
 | |
1J9L
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH VANADATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-28 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SURE PROTEIN FROM T.MARITIMA | | Descriptor: | MAGNESIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, SULFATE ION | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1RHJ
 
 | |
1J9K
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH TUNGSTATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
5S4J
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | Descriptor: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1RHU
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF CASPASE-3 WITH A 5,6,7 TRICYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (3S)-3-[({(2S)-5-[(N-ACETYL-L-ALPHA-ASPARTYL)AMINO]-4-OXO-1,2,4,5,6,7-HEXAHYDROAZEPINO[3,2,1-HI]INDOL-2-YL}CARBONYL)AMINO]-5-(BENZYLSULFANYL)-4-OXOPENTANOIC ACID, Caspase-3 | | Authors: | Becker, J.W, Rotonda, J, Soisson, S.M. | | Deposit date: | 2003-11-14 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Reducing the Peptidyl Features of Caspase-3 Inhibitors: A Structural Analysis.
J.Med.Chem., 47, 2004
|
|
5S4I
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | Descriptor: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4H
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF048 | | Descriptor: | 1-carbamoylpiperidine-4-carboxylic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.175 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4F
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF003 | | Descriptor: | 1,8-naphthyridine, Non-structural protein 3, SULFATE ION | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4G
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF005 | | Descriptor: | Non-structural protein 3, SULFATE ION, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1RHQ
 
 | |
1RHM
 
 | |
1RHK
 
 | |
1RHR
 
 | |
1RHG
 
 | |
2L3H
 
 | | NMR Structure in a Membrane Environment Reveals Putative Amyloidogenic Regions of the SEVI Precursor Peptide PAP248-286 | | Descriptor: | Prostatic acid phosphatase | | Authors: | Ramamoorthy, A, Nanga, R, Brender, J, Vivekanandan, S, Popovych, N. | | Deposit date: | 2010-09-13 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure in a membrane environment reveals putative amyloidogenic regions of the SEVI precursor peptide PAP(248-286).
J.Am.Chem.Soc., 131, 2009
|
|
1GRA
 
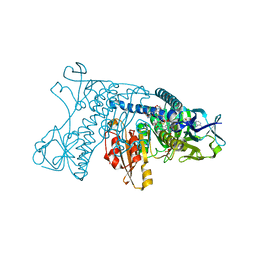 | |
2GRT
 
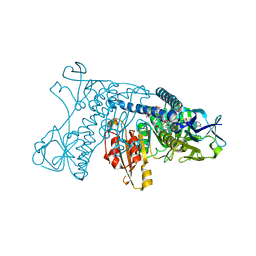 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED GLUTATHIONE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
1GRF
 
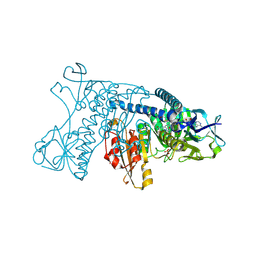 | |
7JTG
 
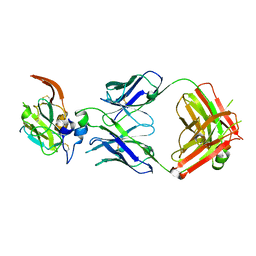 | |
7JTF
 
 | |
5RHD
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with SF013 (Mpro-x2193) | | Descriptor: | 1-[4-(methylsulfonyl)phenyl]piperazine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
