8X26
 
 | | Crystal structure of H5 hemagglutinin from human-infecting H5N8 influenza virus in complex with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Jin, X.Y, Song, H, Han, P, Qi, J.X. | | Deposit date: | 2023-11-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Enhanced binding of Sialyl Lewis X to hemagglutinin contributes to H5N8 mammalian pathogenicity and infectivity
To Be Published
|
|
8X2C
 
 | |
8X2D
 
 | | Crystal structure of H5 hemagglutinin from swan-infecting H5N8 influenza virus complexed with avian receptor analog LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Jin, X.Y, Han, P, Song, H, Qi, J.X. | | Deposit date: | 2023-11-09 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Enhanced binding of Sialyl Lewis X to hemagglutinin contributes to H5N8 mammalian pathogenicity and infectivity
To Be Published
|
|
8X28
 
 | |
8X27
 
 | | Crystal structure of H5 hemagglutinin from human-infecting H5N8 influenza virus in complex with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Jin, X.Y, Han, P, Song, H, Qi, J.X. | | Deposit date: | 2023-11-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Enhanced binding of Sialyl Lewis X to hemagglutinin contributes to H5N8 mammalian pathogenicity and infectivity
To Be Published
|
|
8X29
 
 | | Crystal structure of H5 hemagglutinin from swan-infecting H5N8 influenza virus in complex with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Jin, X.Y, Han, P, Song, H, Qi, J.X. | | Deposit date: | 2023-11-09 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Enhanced binding of Sialyl Lewis X to hemagglutinin contributes to H5N8 mammalian pathogenicity and infectivity
To Be Published
|
|
7WLK
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with Otilonium Bromide(OB) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[diethyl(methyl)-$l^{4}-azanyl]ethyl 4-[(2-octoxyphenyl)carbonylamino]benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLI
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel CaV3.3 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLL
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with pimozide(PMZ) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
8XFD
 
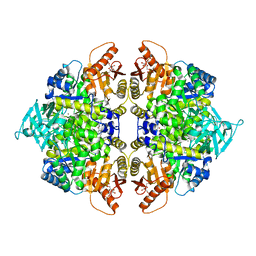 | | Crystal structure of pyruvate kinase tetramer in complex with allosteric activator, Mitapivat (MTPV, AG-348) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Isoform L-type of Pyruvate kinase PKLR, MAGNESIUM ION, ... | | Authors: | Sun, R, Achour, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution Crystal Structure of the Pyruvate Kinase Tetramer in Complex with the Allosteric Activator Mitapivat/AG-348
Crystals, 2024
|
|
8XYR
 
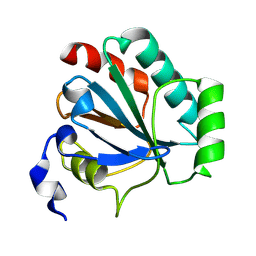 | | De novo designed protein GPX4-2 | | Descriptor: | De novo designed GPX4-2 | | Authors: | Liu, L.J, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYV
 
 | | De novo designed protein 0705-5 | | Descriptor: | De novo designed protein 0705-5 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYW
 
 | | De novo designed protein Trx-3 | | Descriptor: | De novo designed Trx-3 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYU
 
 | | De novo designed protein GPX4-3 | | Descriptor: | De novo designed GPX4-3 | | Authors: | Guo, Z, Liu, J.L, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYS
 
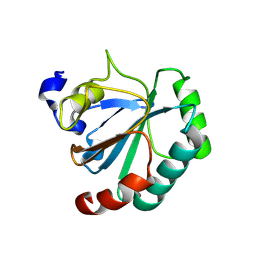 | | De novo designed protein GPX4-1 | | Descriptor: | De novo designed GPX4-1 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYT
 
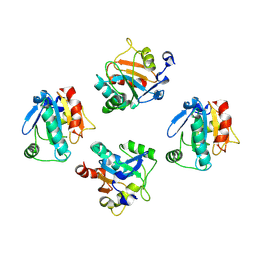 | | De novo designed protein GPX4-4 | | Descriptor: | De novo designed GPX4-4 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7WK5
 
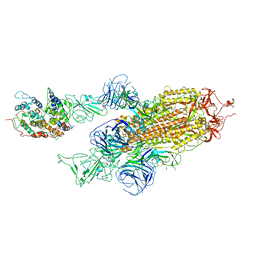 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Han, W.Y, Wang, Y.F. | | Deposit date: | 2022-01-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
7WK3
 
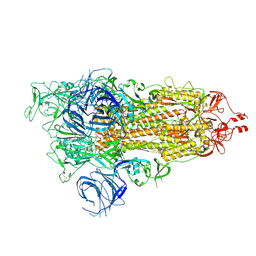 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
7VOP
 
 | | Cryo-EM structure of Xenopus laevis nuclear pore complex cytoplasmic ring subunit | | Descriptor: | GATOR complex protein SEC13, IL4I1 protein, MGC154553 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7WLJ
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with mibefradil (MIB) | | Descriptor: | (1S,2S)-2-(2-{[3-(1H-benzimidazol-2-yl)propyl](methyl)amino}ethyl)-6-fluoro-1-(propan-2-yl)-1,2,3,4-tetrahydronaphthalen-2-yl methoxyacetate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7C91
 
 | | Blasnase-T13A with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7CBU
 
 | | Blasnase-T13A with L-Asp | | Descriptor: | ASPARTIC ACID, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7CB4
 
 | | Crystal structures of of BlAsnase | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7C8X
 
 | | Blasnase-T13A with L-asn | | Descriptor: | ASPARAGINE, Asparaginase, FORMIC ACID, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7CBR
 
 | | Blasnase-T13A with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-13 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
