1VI9
 
 | | Crystal structure of pyridoxamine kinase | | Descriptor: | BETA-MERCAPTOETHANOL, Pyridoxamine kinase, SULFATE ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VH6
 
 | | Crystal structure of a flagellar protein | | Descriptor: | Flagellar protein fliS | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHK
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yqeU | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
3NMI
 
 | | Crystal structure of the phenanthroline-modified cytochrome cb562 variant, MBP-Phen2 | | Descriptor: | ACETATE ION, N-1,10-phenanthrolin-5-ylacetamide, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Radford, R.J, Tezcan, F.A. | | Deposit date: | 2010-06-22 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Porous protein frameworks with unsaturated metal centers in sterically encumbered coordination sites.
Chem.Commun.(Camb.), 47, 2011
|
|
3NSH
 
 | | BACE-1 in complex with ELN475957 | | Descriptor: | Beta-secretase 1, N-[(1S,2R)-1-(3,5-difluorobenzyl)-3-({1-[4-(2,2-dimethylpropyl)thiophen-2-yl]cyclopropyl}amino)-2-hydroxypropyl]acetamide | | Authors: | Probst, G.D, Bowers, S, Sealy, J.M, Brecht, E, Yao, N. | | Deposit date: | 2010-07-01 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of hydroxyethylamine (HEA) BACE-1 inhibitors: structure-activity relationship of the aryl region.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1S8E
 
 | | Crystal structure of Mre11-3 | | Descriptor: | MANGANESE (II) ION, exonuclease putative | | Authors: | Hopfner, K.P. | | Deposit date: | 2004-02-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of Mre11-3
Nucleic Acids Res., 32, 2004
|
|
3NSJ
 
 | | The X-ray crystal structure of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Law, R.H, Whisstock, J.C, Caradoc-Davies, T.T. | | Deposit date: | 2010-07-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structural basis for membrane binding and pore formation by lymphocyte perforin.
Nature, 468, 2010
|
|
3NWW
 
 | | P38 Alpha kinase complexed with a 2-aminothiazol-5-yl-pyrimidine based inhibitor | | Descriptor: | 1-[2-(2-{[2-(dimethylamino)ethyl]amino}-6-{2-[(1-methylethyl)amino]-1,3-thiazol-5-yl}pyrimidin-4-yl)benzyl]-3-ethylurea, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-07-12 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Utilization of a nitrogen-sulfur nonbonding interaction in the design of new 2-aminothiazol-5-yl-pyrimidines as p38alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1ICA
 
 | |
1I6B
 
 | |
3Q4F
 
 | | Crystal structure of xrcc4/xlf-cernunnos complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Ropars, V, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2010-12-23 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1I1Y
 
 | |
3Q0D
 
 | | Crystal structure of SUVH5 SRA- hemi methylated CG DNA complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*(5CM)P*GP*TP*CP*AP*G)-3'), ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3704 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
1VES
 
 | | Structure of New Antigen Receptor variable domain from sharks | | Descriptor: | New Antigen Receptor variable domain | | Authors: | Streltsov, V.A. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VH2
 
 | |
1VH9
 
 | | Crystal structure of a putative thioesterase | | Descriptor: | Hypothetical protein ybdB | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1HSI
 
 | |
1HSG
 
 | |
1HUN
 
 | |
3OX2
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | 2-hydroxy-8,9-dimethoxy-6H-isoindolo[2,1-a]indol-6-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
1T93
 
 | | Evidence for Multiple Substrate Recognition and Molecular Mechanism of C-C reaction by Cytochrome P450 CYP158A2 from Streptomyces Coelicolor A3(2) | | Descriptor: | FLAVIOLIN, PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Zhao, B, Sundaramoorthy, M, Waterman, M.R. | | Deposit date: | 2004-05-14 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2.
J.Biol.Chem., 280, 2005
|
|
1GG2
 
 | |
1WBI
 
 | | AVR2 | | Descriptor: | AVIDIN-RELATED PROTEIN 2, BIOTIN, GLYCEROL, ... | | Authors: | Airenne, T.T, Hytonen, V.P, Maatta, J.H, Kidron, H, Halling, K.K, Horha, J, Kulomaa, T, Nyholm, T.K.M, Johnson, M.S, Salminen, T.A, Kulomaa, M.S. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Avidin Related Protein 2 Shows Unique Structural and Functional Features Among the Avidin Protein Family.
Bmc Biotechnol., 5, 2005
|
|
3PJZ
 
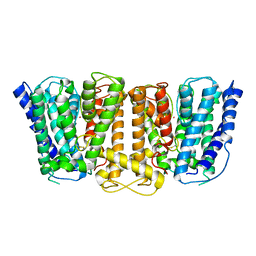 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
3MVM
 
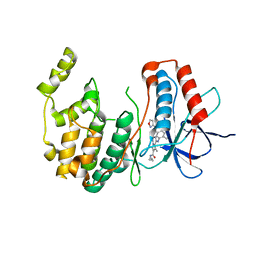 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7V | | Descriptor: | 4-{[5-(isoxazol-3-ylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38? Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
