1UO9
 
 | | Deacetoxycephalosporin C synthase complexed with succinate | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, SUCCINIC ACID | | Authors: | Valegard, K, Terwisscha Van scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UOB
 
 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and penicillin G | | Descriptor: | 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UNB
 
 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and ampicillin | | Descriptor: | (2S,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-09 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UOG
 
 | | Deacetoxycephalosporin C synthase complexed with deacetoxycephalosporin C | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, DEACETOXYCEPHALOSPORIN-C, FE (II) ION | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-17 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
6ITX
 
 | |
6JDS
 
 | |
6JDU
 
 | | Crystal structure of PRRSV nsp10 (helicase) | | Descriptor: | CALCIUM ION, PP1b, ZINC ION | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2019-02-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
2MJV
 
 | |
6JDR
 
 | |
4K18
 
 | | Structure of PIM-1 kinase bound to 5-(4-cyanobenzyl)-N-(4-fluorophenyl)-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | 5-(4-cyanobenzyl)-N-(4-fluorophenyl)-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H, Steffek, M. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Discovery of novel pyrazolo[1,5-a]pyrimidines as potent pan-Pim inhibitors by structure- and property-based drug design.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4K0Y
 
 | | Structure of PIM-1 kinase bound to N-(4-fluorophenyl)-7-hydroxy-5-(piperidin-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | N-(4-fluorophenyl)-7-hydroxy-5-(piperidin-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H, Steffek, M. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Discovery of novel pyrazolo[1,5-a]pyrimidines as potent pan-Pim inhibitors by structure- and property-based drug design.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3O2G
 
 | | Crystal Structure of Human gamma-butyrobetaine,2-oxoglutarate dioxygenase 1 (BBOX1) | | Descriptor: | 1,2-ETHANEDIOL, 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, Gamma-butyrobetaine dioxygenase, ... | | Authors: | Krojer, T, Kochan, G, McDonough, M.A, von Delft, F, Leung, I.K.H, Henry, L, Claridge, T.D.W, Pilka, E, Ugochukwu, E, Muniz, J, Filippakopoulos, P, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and mechanistic studies on gamma-butyrobetaine hydroxylase.
Chem. Biol., 17, 2010
|
|
5EI3
 
 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | Eukaryotic translation initiation factor 4 gamma, Eukaryotic translation initiation factor 4E, SULFATE ION, ... | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-29 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
5EKV
 
 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | 3-[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-4-oxidanyl-cyclobut-3-ene-1,2-dione, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
5EHC
 
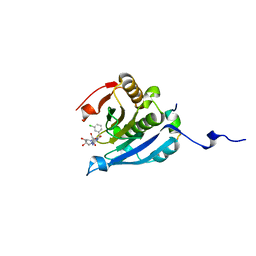 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | 3-[[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-chlorophenyl)methyl]-6-oxidanylidene-1~{H}-purin-7-ium-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-4-oxidanyl-cyclobut-3-ene-1,2-dione, Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
1Z3W
 
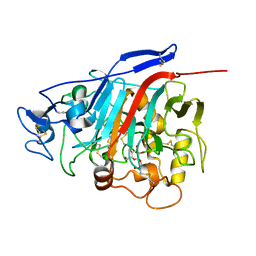 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with cellobioimidazole | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Vasella, A, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
1Z3T
 
 | |
3FAA
 
 | |
3HA4
 
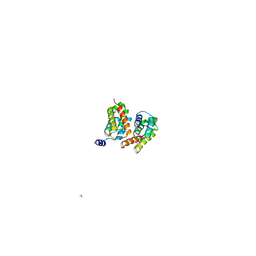 | |
2XDV
 
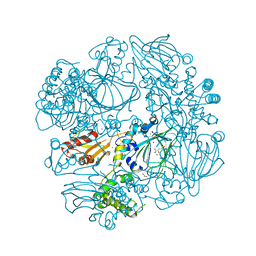 | | Crystal Structure of the Catalytic Domain of FLJ14393 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, MANGANESE (II) ION, ... | | Authors: | Krojer, T, Muniz, J.R.C, Ng, S.S, Pilka, E, Guo, K, Pike, A.C.W, Filippakopoulos, P, Knapp, S, Kavanagh, K.L, Gileadi, O, Bunkoczi, G, Yue, W.W, Niesen, F, Sobott, F, Fedorov, O, Savitsky, P, Kochan, G, Daniel, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2010-05-07 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
6DXO
 
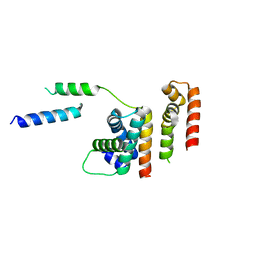 | | 1.8 A structure of RsbN-BldN complex. | | Descriptor: | BldN, RNA polymerase ECF-subfamily sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the RsbN-sigma BldN complex from Streptomyces venezuelae defines a new structural class of anti-sigma factor.
Nucleic Acids Res., 46, 2018
|
|
5EIR
 
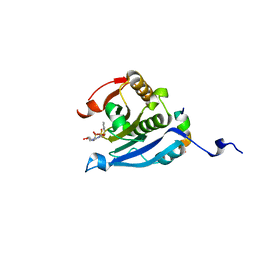 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E, SULFATE ION, ... | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
2JFD
 
 | | Structure of the MAT domain of human FAS | | Descriptor: | FATTY ACID SYNTHASE | | Authors: | Bunkoczi, G, Kavanagh, K, Hozjan, V, Rojkova, A, Wu, X, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Smith, S, Oppermann, U. | | Deposit date: | 2007-01-31 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for different specificities of acyltransferases associated with the human cytosolic and mitochondrial fatty acid synthases.
Chem. Biol., 16, 2009
|
|
4K1B
 
 | | Structure of PIM-1 kinase bound to N-(5-(2-fluorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-((((3R,4R)-3-fluoropiperidin-4-yl)methyl)amino)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | N-[5-(2-fluorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]-5-({[(3R,4R)-3-fluoropiperidin-4-yl]methyl}amino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H, Steffek, M. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Discovery of novel pyrazolo[1,5-a]pyrimidines as potent pan-Pim inhibitors by structure- and property-based drug design.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1GFW
 
 | |
