3BUS
 
 | | Crystal Structure of RebM | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCoy, J.G, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of the rebeccamycin sugar 4'-O-methyltransferase RebM.
J.Biol.Chem., 283, 2008
|
|
5F9P
 
 | | Crystal structure study of anthrone oxidase-like protein | | Descriptor: | Anthrone oxidase-like protein, GLYCEROL | | Authors: | Gao, X, Wu, D, Fan, K, Liu, Z.-J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Structure and Function of a C-C Bond Cleaving Oxygenase in Atypical Angucycline Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
7D5G
 
 | |
8JA0
 
 | | Cryo-EM structure of the NmeCas9-sgRNA-AcrIIC4 ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (117-MER), Uncharacterized protein | | Authors: | Yin, H, Li, Z, Yu, G.M, Li, X.Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Inhibitory mechanism of CRISPR-Cas9 by AcrIIC4.
Nucleic Acids Res., 51, 2023
|
|
8HZQ
 
 | | Bacillus subtilis SepF protein assembly (wild type) | | Descriptor: | Cell division protein SepF | | Authors: | Liu, W. | | Deposit date: | 2023-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular basis for curvature formation in SepF polymerization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8HZT
 
 | |
7WMJ
 
 | | A novel chemical derivative(71) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(phenylcarbonyl)phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMO
 
 | | A novel chemical derivative(92) of THRB agonist | | Descriptor: | 2-[[1-ethoxy-7-[4-(3-fluoranyl-5-methoxy-phenyl)carbonyl-2,6-dimethyl-phenoxy]-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMH
 
 | | A novel chemical derivative(56) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenylphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMG
 
 | | A novel chemical derivative(52) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenoxyphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMN
 
 | | A novel chemical derivative(89) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(3-methylphenyl)carbonyl-phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WML
 
 | | A novel chemical derivative(85) of THRB agonist | | Descriptor: | 2-[[7-[4-(3,5-dimethylphenyl)carbonyl-2,6-dimethyl-phenoxy]-1-ethoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WLX
 
 | | A novel chemical derivative(53) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-[4-(phenylmethyl)phenoxy]isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
2L65
 
 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
7N9T
 
 | | CryoEM structure of SARS-CoV-2 Spike in complex with Nb17 | | Descriptor: | Nanobody Nb17, Spike glycoprotein | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2021-06-18 | | Release date: | 2021-08-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes
Nat Commun, 12, 2021
|
|
7MTQ
 
 | | CryoEM Structure of Full-Length mGlu2 in Inactive-State Bound to Antagonist LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Seven, A.B, Barros-Alvarez, X, Skiniotis, G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | G-protein activation by a metabotropic glutamate receptor.
Nature, 595, 2021
|
|
7MTR
 
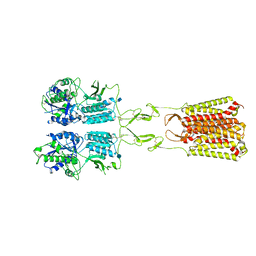 | | CryoEM Structure of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and Glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethyl)thieno[2,3-d]pyrimidin-4-amine, GLUTAMIC ACID, ... | | Authors: | Seven, A.B, Barros-Alvarez, X, Skiniotis, G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | G-protein activation by a metabotropic glutamate receptor.
Nature, 595, 2021
|
|
7MTS
 
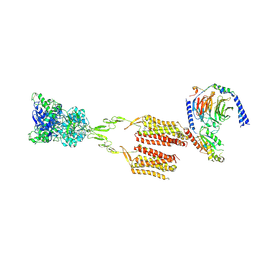 | | CryoEM Structure of mGlu2 - Gi Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethyl)thieno[2,3-d]pyrimidin-4-amine, GLUTAMIC ACID, ... | | Authors: | Seven, A.B, Barros-Alvarez, X, Skiniotis, G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | G-protein activation by a metabotropic glutamate receptor.
Nature, 595, 2021
|
|
7MEJ
 
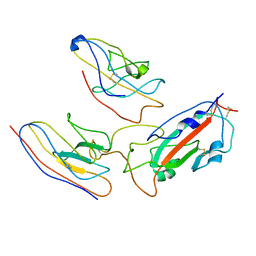 | |
7MDW
 
 | |
7ME7
 
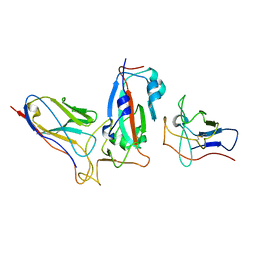 | |
9IM5
 
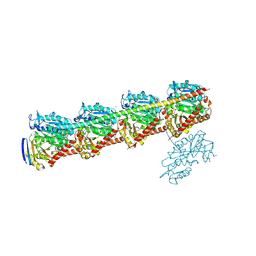 | | Tubulin-RB3(MUT)-TTL-Y12 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-02 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IMO
 
 | | Crystal structure of Tubulin-RB3-TTL-Y12 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-04 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9JSP
 
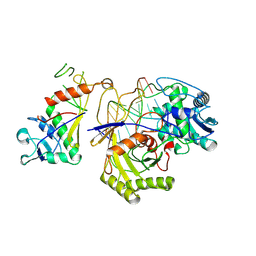 | | inactive NbaSPARDA complexes | | Descriptor: | Ago, DNA (5'-D(P*GP*CP*TP*GP*TP*GP*CP*AP*GP*TP*AP*TP*T)-3'), DREN-APAZ, ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-09-30 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 35, 2025
|
|
9JSZ
 
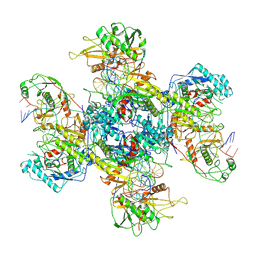 | | active NbaSPARDA complexes | | Descriptor: | Ago, DNA (5'-D(*TP*AP*TP*CP*GP*TP*CP*AP*GP*CP*TP*GP*TP*GP*CP*AP*GP*TP*AP*TP*T)-3'), DREN-APAZ, ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-10-01 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 35, 2025
|
|
