4R1V
 
 | | Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitors | | Descriptor: | 3-[1-(3-{5-[(1-methylpiperidin-4-yl)methoxy]pyrimidin-2-yl}benzyl)-6-oxo-1,6-dihydropyridazin-3-yl]benzonitrile, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Blaukat, A, Bladt, F, Friese-Hamim, M, Knuehl, C, Fittschen, C, Graedler, U, Meyring, M, Dorsch, D, Stieber, F, Schadt, O. | | Deposit date: | 2014-08-07 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7LZE
 
 | | Cryo-EM Structure of disulfide stabilized HMPV F v4-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8VSG
 
 | | SARS-CoV-2 main protease with covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-(1-phenylcyclopropane-1-carbonyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bell, J.A, Bandera, A.M. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
8VQX
 
 | | Structure of SARS-CoV-2 main protease with potent peptide aldehyde inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2024-01-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
8TE7
 
 | | Structure of TRNM-f.01 | | Descriptor: | TRNM-f.01 Fab Heavy Chain, TRNM-f.01 Fab Light Chain | | Authors: | Bender, M.F, Olia, A.S, Kwong, P.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TJR
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-a.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody HERH-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TJS
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO GPZ6-a.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody GPZ6-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Becker, J.E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TDX
 
 | | TRNM-b.01 in complex with HIV Env fusion peptide | | Descriptor: | Env Fusion Peptide, TRNM-b.01 Fab Heavy Chain, TRNM-b.01 Fab Light Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TNU
 
 | | Cryo-EM structure of TRNM-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TOP
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with antibody GPZ6-b.01 targeting the fusion peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP glycoprotein gp41, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TO7
 
 | | Cryo-EM structure of HERH-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HERH-b*01 heavy chain, ... | | Authors: | Roark, R.S, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TO9
 
 | | Cryo-EM structure of TRNM-f*01 Fab in complex with HIV-1 Env trimer ConC SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL2
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-c.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL3
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-d.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP glycoprotein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL5
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-c.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TKC
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-b.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL4
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-e.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
4R1Y
 
 | | Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitor | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3-(diethylamino)propyl (3-{[5-(3,4-dimethoxyphenyl)-2-oxo-2H-1,3,4-thiadiazin-3(6H)-yl]methyl}phenyl)carbamate, Hepatocyte growth factor receptor | | Authors: | Blaukat, A, Bladt, F, Friese-Hamim, M, Knuehl, C, Fittschen, C, Graedler, U, Meyring, M, Dorsch, D, Stieber, F, Schadt, O. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8F7T
 
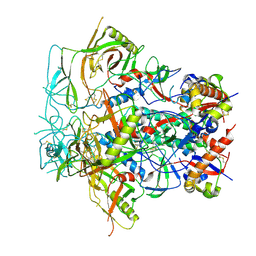 | | Glycan-Base ConC Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env gp120, ... | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-20 | | Release date: | 2023-09-27 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases.
Iscience, 26, 2023
|
|
8G4M
 
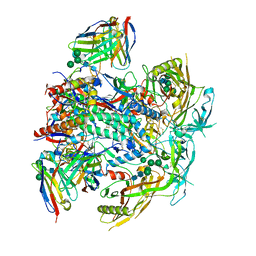 | | Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Wang, S, Morano, N.C, Shapiro, L, Kwong, P.D. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Cell Rep, 42, 2023
|
|
8G4T
 
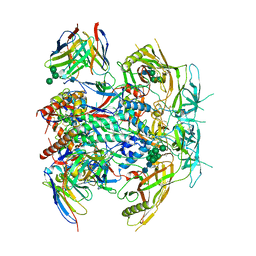 | | Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein BG505 DS-SOSIP gp120, ... | | Authors: | Wang, S, Kwong, P.D. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Cell Rep, 42, 2023
|
|
4QM0
 
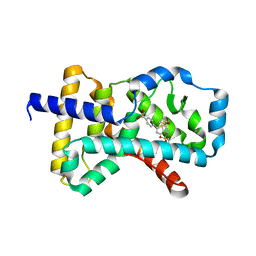 | | Crystal structure of RORc in complex with a tertiary sulfonamide inverse agonist | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-methylpropyl)-N-({5-[4-(methylsulfonyl)phenyl]thiophen-2-yl}methyl)-1-phenylmethanesulfonamide, Nuclear receptor ROR-gamma | | Authors: | Boenig, G, Hymowitz, S.G, Wang, W. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Reduction in lipophilicity improved the solubility, plasma-protein binding, and permeability of tertiary sulfonamide RORc inverse agonists.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8GY0
 
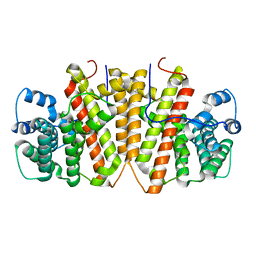 | | Agrocybe pediades linalool sunthase (Ap.LS) | | Descriptor: | Terpene synthase | | Authors: | Rehka, T, Sharma, D, Lin, F, Lim, C, Choong, Y.K, Chacko, J, Zhang, C. | | Deposit date: | 2022-09-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural Understanding of Fungal Terpene Synthases for the Formation of Linear or Cyclic Terpene Products.
Acs Catalysis, 13, 2023
|
|
8VW5
 
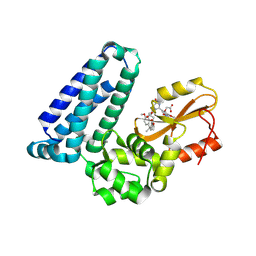 | | Crystal structure of Cbl-b TKB bound to compound 2 | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL-B, MAGNESIUM ION, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
8VW4
 
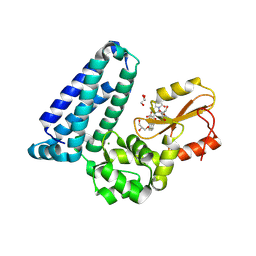 | | Crystal structure of Cbl-b TKB bound to compound 26 | | Descriptor: | (7-methoxy-2-{2-[(1S,3S,4S)-3-(3-methoxy-2-methyl-5-nitrophenyl)-1-methyl-5-oxo-1,5-dihydroimidazo[1,5-a]pyridin-2(3H)-yl]-2-oxoethoxy}quinolin-8-yl)acetic acid, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
