7E9Q
 
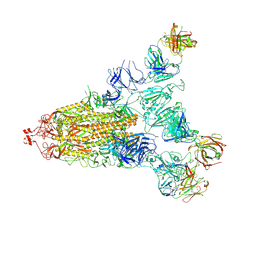 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9P
 
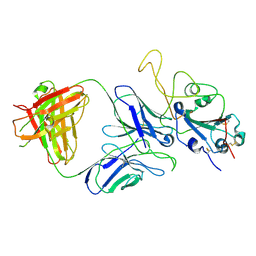 | |
4RBZ
 
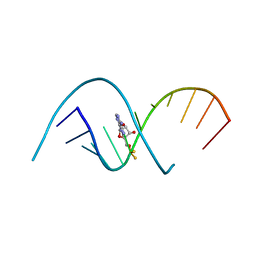 | |
7ENG
 
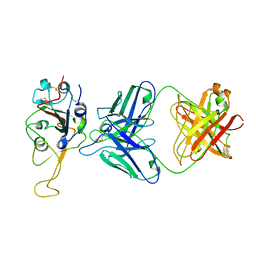 | |
7E9N
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7VVE
 
 | | Complex structure of a leaf-branch compost cutinase variant in complex with mono(2-hydroxyethyl) terephthalic acid | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W44
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7VVC
 
 | | Crystal structure of inactive mutant of leaf-branch compost cutinase variant | | Descriptor: | ACETATE ION, ACETIC ACID, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | Descriptor: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
8JA8
 
 | |
8JAC
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-16 | | Descriptor: | N-[[(2R,3S,4S,5R,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl]ethanamide, SULFATE ION, Trehalose-binding lipoprotein LpqY, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAB
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-06 | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{R})-2-(fluoranylmethyl)-6-[(2~{R},3~{R},4~{S},5~{S},6~{S})-6-(fluoranylmethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-oxane-3,4,5-triol, SULFATE ION, Trehalose-binding lipoprotein LpqY | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAD
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-17 | | Descriptor: | BENZOIC ACID, SULFATE ION, Trehalose-binding lipoprotein LpqY, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JA7
 
 | | Cryo-EM structure of Mycobacterium tuberculosis LpqY-SugABC in complex with trehalose | | Descriptor: | Trehalose import ATP-binding protein SugC, Trehalose transport system permease protein SugA, Trehalose transport system permease protein SugB, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JA9
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-03 | | Descriptor: | SULFATE ION, Trehalose-binding lipoprotein LpqY, alpha-D-glucopyranose-(1-1)-(2~{S},3~{R},4~{S},5~{S},6~{S})-6-[(2-azanylhydrazinyl)methyl]oxane-2,3,4,5-tetrol | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAA
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-04 | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{S})-6-[(2-azanylhydrazinyl)methyl]oxane-2,3,4,5-tetrol, SULFATE ION, Trehalose-binding lipoprotein LpqY | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YG1
 
 | | Cryo-EM structure of the C-terminal domain of the human sodium-chloride cotransporter | | Descriptor: | Solute carrier family 12 member 3 | | Authors: | Nan, J, Yang, X.M, Shan, Z.Y, Yuan, Y.F, Zhang, Y.Q. | | Deposit date: | 2022-07-09 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structure of the human sodium-chloride cotransporter NCC.
Sci Adv, 8, 2022
|
|
7YG0
 
 | | Cryo-EM structure of human sodium-chloride cotransporter | | Descriptor: | Solute carrier family 12 member 3 | | Authors: | Nan, J, Yang, X.M, Shan, Z.Y, Yuan, Y.F, Zhang, Y.Q. | | Deposit date: | 2022-07-09 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structure of the human sodium-chloride cotransporter NCC.
Sci Adv, 8, 2022
|
|
6NZR
 
 | |
4GA7
 
 | | Crystal structure of human serpinB1 mutant | | Descriptor: | Leukocyte elastase inhibitor | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4GAW
 
 | | Crystal structure of active human granzyme H | | Descriptor: | CHLORIDE ION, Granzyme H, SULFATE ION | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
6NZF
 
 | |
6NZE
 
 | |
