8DVR
 
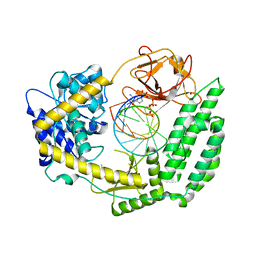 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+AMPPNP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, GUANOSINE-5'-TRIPHOSPHATE, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVS
 
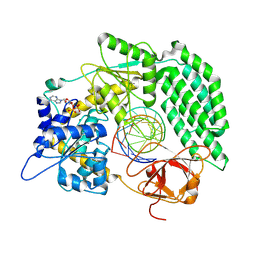 | | Cryo-EM structure of RIG-I bound to the end of OHSLR30 (+ATP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, MAGNESIUM ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNY
 
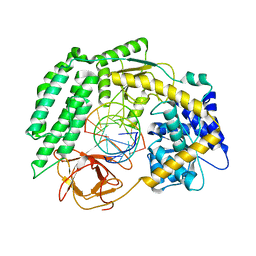 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVU
 
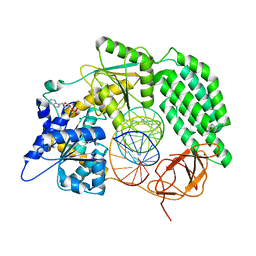 | |
1MRZ
 
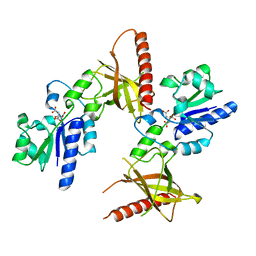 | | Crystal structure of a flavin binding protein from Thermotoga Maritima, TM379 | | Descriptor: | CITRIC ACID, Riboflavin kinase/FMN adenylyltransferase | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-09-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a flavin-binding protein from Thermotoga Maritima
Proteins, 52, 2003
|
|
1PWJ
 
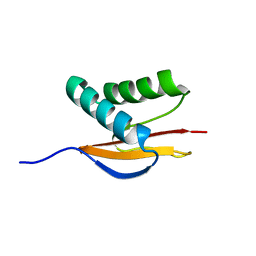 | | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of Domain Swapped Dimer Assembly | | Descriptor: | dynein light chain-2 | | Authors: | Wang, W, Lo, K.W.-H, Kan, H.-M, Fan, J.-S, Zhang, M. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of the Domain-swapped Dimer Assembly
J.Biol.Chem., 278, 2003
|
|
1PWK
 
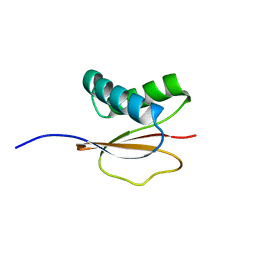 | | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of Domain Swapped Dimer Assembly | | Descriptor: | dynein light chain-2 | | Authors: | Wang, W, Lo, K.W.-H, Kan, H.-M, Fan, J.-S, Zhang, M. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of the Domain-swapped Dimer Assembly
J.Biol.Chem., 278, 2003
|
|
4LJR
 
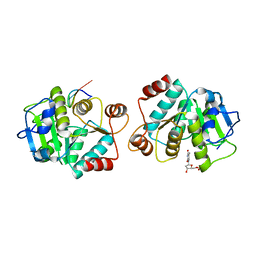 | |
4LJL
 
 | |
4LJK
 
 | |
8G7U
 
 | |
8G7T
 
 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-end) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
8G7V
 
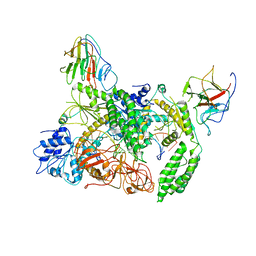 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
1ZDU
 
 | | The Crystal Structure of Human Liver Receptor Homologue-1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2, ... | | Authors: | Wang, W, Zhang, C, Marimuthu, A, Krupka, H.I, Tabrizizad, M, Shelloe, R, Mehra, U, Eng, K, Nguyen, H, Settachatgul, C, Powell, B, Milburn, M.V, West, B.L. | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ZDT
 
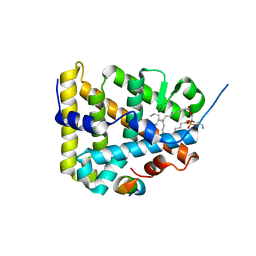 | | The Crystal Structure of Human Steroidogenic Factor-1 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, Steroidogenic factor 1 | | Authors: | Wang, W, Zhang, C, Marimuthu, A, Krupka, H.I, Tabrizizad, M, Shelloe, R, Mehra, U, Eng, K, Nguyen, H, Settachatgul, C, Powell, B, Milburn, M.V, West, B.L. | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6WPG
 
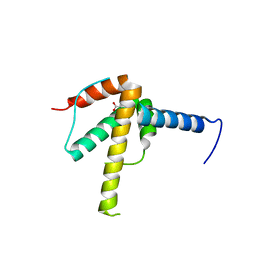 | | Structural Basis of Salicylic Acid Perception by Arabidopsis NPR Proteins | | Descriptor: | 2-HYDROXYBENZOIC ACID, Regulatory protein NPR4 | | Authors: | Wang, W, Withers, J, Li, H, Zwack, P.J, Rusnac, D.V, Shi, H, Liu, L, Yan, S, Hinds, T.R, Guttman, M, Dong, X, Zheng, N. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Structural basis of salicylic acid perception by Arabidopsis NPR proteins.
Nature, 586, 2020
|
|
3PX0
 
 | |
6NTV
 
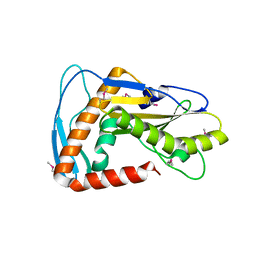 | | SFTSV L endonuclease domain | | Descriptor: | RNA polymerase | | Authors: | Wang, W, Amarasinghe, G.K. | | Deposit date: | 2019-01-30 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cap-Snatching SFTSV Endonuclease Domain Is an Antiviral Target.
Cell Rep, 30, 2020
|
|
5XGQ
 
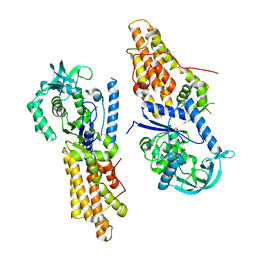 | |
5XET
 
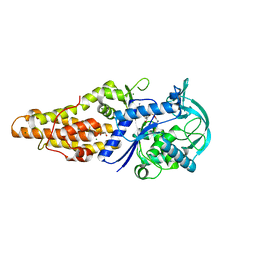 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
5Z1Q
 
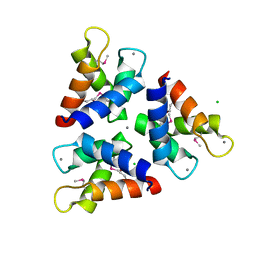 | | Crystal structures of the trimeric N-terminal domain of Ciliate Euplotes octocarinatus Centrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Centrin protein | | Authors: | Wang, W, Zhao, Y, Yang, B, Wang, H. | | Deposit date: | 2017-12-27 | | Release date: | 2018-04-25 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the trimeric N-terminal domain of ciliate Euplotes octocarinatus centrin binding with calcium ions
Protein Sci., 27, 2018
|
|
6CU1
 
 | | X-ray structure of the S. typhimurium YrlA effector-binding module | | Descriptor: | MAGNESIUM ION, SULFATE ION, YrlA effector-binding module | | Authors: | Wang, W, Chen, X, Wolin, S.L, Xiong, Y. | | Deposit date: | 2018-03-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for tRNA Mimicry by a Bacterial Y RNA.
Structure, 26, 2018
|
|
3PX6
 
 | |
3PV8
 
 | |
4FXW
 
 | | Structure of phosphorylated SF1 complex with U2AF65-UHM domain | | Descriptor: | SULFATE ION, Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Wang, W, Bauer, W.J, Wedekind, J.E, Kielkopf, C.L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-01-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
