3CGI
 
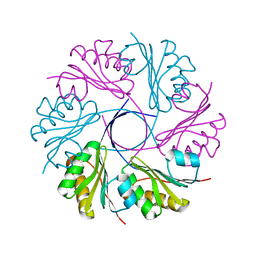 | |
1MY6
 
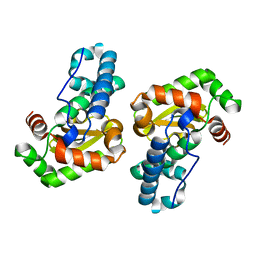 | | The 1.6 A Structure of Fe-Superoxide Dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus : Correlation of EPR and Structural Characteristics | | Descriptor: | FE (III) ION, Iron (III) Superoxide Dismutase | | Authors: | Yoshida, S.M, Cascio, D, Sawaya, M.R, Yeates, T.O, Kerfeld, C.A. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A resolution structure of Fe-superoxide dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus.
J.BIOL.INORG.CHEM., 8, 2003
|
|
1MZ4
 
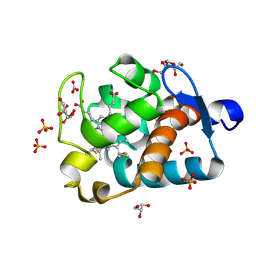 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
3CJB
 
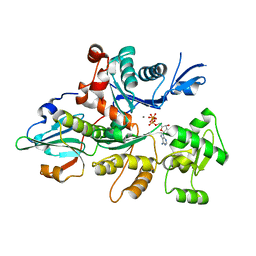 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with Gelsolin-segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CIM
 
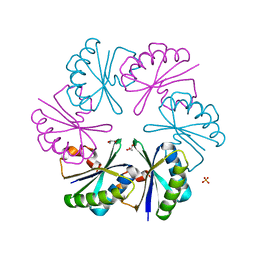 | | Carboxysome shell protein, CcmK2 C-terminal deletion mutant | | Descriptor: | Carbon dioxide-concentrating mechanism protein ccmK homolog 2, GLYCEROL, SULFATE ION | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2008-03-11 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Insights from multiple structures of the shell proteins from the beta-carboxysome.
Protein Sci., 18, 2009
|
|
1F1F
 
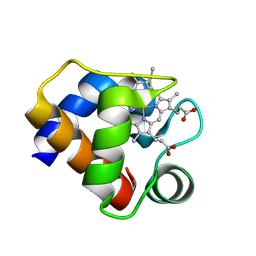 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
3DN9
 
 | |
1KR5
 
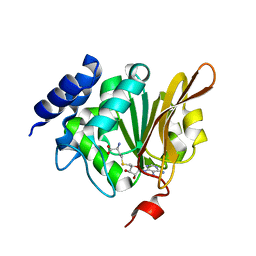 | | Crystal structure of human L-isoaspartyl methyltransferase | | Descriptor: | Protein-L-isoaspartate O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ryttersgaard, C, Griffith, S.C, Sawaya, M.R, MacLaren, D.C, Clarke, S, Yeates, T.O. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human L-isoaspartyl methyltransferase.
J.Biol.Chem., 277, 2002
|
|
3BN4
 
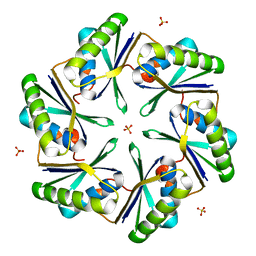 | | Carboxysome Subunit, CcmK1 | | Descriptor: | Carbon dioxide-concentrating mechanism protein ccmK homolog 1, SULFATE ION | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
2Q31
 
 | | Actin Dimer Cross-linked Between Residues 41 and 374 and proteolytically cleaved by subtilisin between residues 47 and 48. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Pashkov, I, Kudryashov, D.S, Reisler, E, Yeates, T.O. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple crystal structures of actin dimers and their implications for interactions in the actin filament.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1L2H
 
 | | Crystal structure of Interleukin 1-beta F42W/W120F mutant | | Descriptor: | Interleukin 1-beta | | Authors: | Rudolph, M.G, Kelker, M.S, Schneider, T.R, Yeates, T.O, Oseroff, V, Heidary, D.K, Jennings, P.A, Wilson, I.A. | | Deposit date: | 2002-02-21 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Use of multiple anomalous dispersion to phase highly merohedrally twinned crystals of interleukin-1beta.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2PLT
 
 | |
3CJC
 
 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with DNase I and Gelsolin-segment 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3DNC
 
 | | Carboxysome shell protein, CcmK2 C-terminal deletion mutant, with a closer spacing between hexamers | | Descriptor: | Carbon dioxide-concentrating mechanism protein ccmK homolog 2, GLYCEROL, SULFATE ION | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2008-07-01 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights from multiple structures of the shell proteins from the beta-carboxysome.
Protein Sci., 18, 2009
|
|
1JG1
 
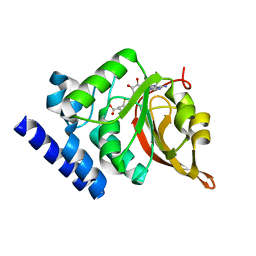 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG4
 
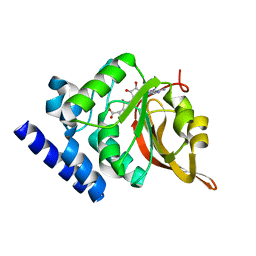 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG3
 
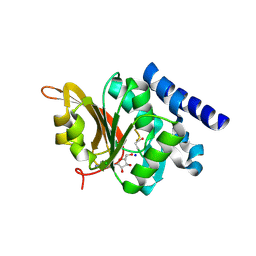 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | Descriptor: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
3HIP
 
 | |
4TME
 
 | |
4TLH
 
 | |
4TM6
 
 | |
4U6I
 
 | | Crystal Structure of the EutL Microcompartment Shell Protein from Clostridium Perfringens Bound to Vitamin B12 | | Descriptor: | COBALAMIN, Ethanolamine utilization protein EutL, SODIUM ION | | Authors: | Thompson, M.C, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4W7F
 
 | |
4ZSV
 
 | |
4ZSZ
 
 | |
