7B7J
 
 | |
1JCM
 
 | | TRPC STABILITY MUTANT CONTAINING AN ENGINEERED DISULPHIDE BRIDGE AND IN COMPLEX WITH A CDRP-RELATED SUBSTRATE | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, INDOLE-3-GLYCEROL-PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Ivens, A, Mayans, O, Szadkowski, H, Wilmanns, M, Kirschner, K. | | Deposit date: | 2001-06-10 | | Release date: | 2002-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stabilization of a (betaalpha)8-barrel protein by an engineered disulfide bridge.
Eur.J.Biochem., 269, 2002
|
|
2Y25
 
 | |
2Y23
 
 | |
1G2B
 
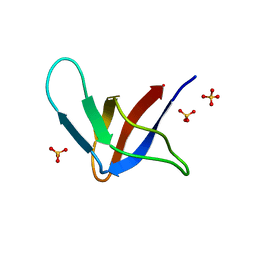 | | ALPHA-SPECTRIN SRC HOMOLOGY 3 DOMAIN, CIRCULAR PERMUTANT, CUT AT N47-D48 | | Descriptor: | SPECTRIN ALPHA CHAIN, SULFATE ION | | Authors: | Berisio, R, Viguera, A.R, Serrano, L, Wilmanns, M. | | Deposit date: | 2000-10-18 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic resolution structure of a mutant of the spectrin SH3 domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1BBZ
 
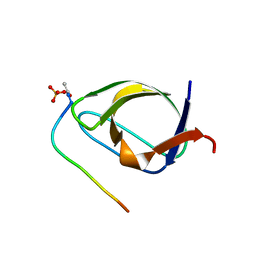 | |
4OP0
 
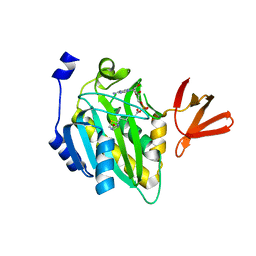 | | Crystal structure of biotin protein ligase (RV3279C) of Mycobacterium tuberculosis, complexed with biotinyl-5'-AMP | | Descriptor: | BIOTINYL-5-AMP, BirA bifunctional protein, SULFATE ION | | Authors: | Ma, Q, Wilmanns, M, Akhter, Y. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active site conformational changes upon reaction intermediate biotinyl-5'-AMP binding in biotin protein ligase from Mycobacterium tuberculosis.
Protein Sci., 23, 2014
|
|
3ZR4
 
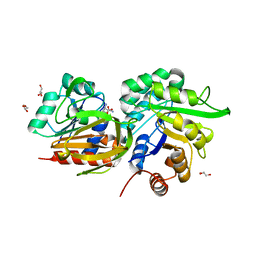 | | STRUCTURAL EVIDENCE FOR AMMONIA TUNNELING ACROSS THE (BETA-ALPHA)8 BARREL OF THE IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE BIENZYME COMPLEX | | Descriptor: | GLUTAMINE, GLYCEROL, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF, ... | | Authors: | Vega, M.C, Kuper, J, Haeger, M.C, Mohrlueder, J, Marquardt, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2011-06-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
6ZVA
 
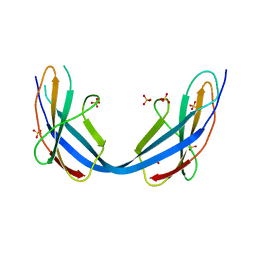 | | Crystal structure of My5 | | Descriptor: | Myomesin-1, SULFATE ION | | Authors: | Sauer, F, Wilmanns, M. | | Deposit date: | 2020-07-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Myomesin-1
To Be Published
|
|
1K9V
 
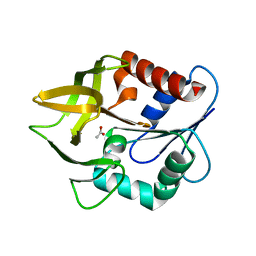 | | Structural evidence for ammonia tunelling across the (beta-alpha)8-barrel of the imidazole glycerol phosphate synthase bienzyme complex | | Descriptor: | ACETIC ACID, Amidotransferase hisH | | Authors: | Douangamath, A, Walker, M, Beismann-Driemeyer, S, Vega-Fernandez, M.C, Sterner, R, Wilmanns, M. | | Deposit date: | 2001-10-31 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex.
Structure, 10, 2002
|
|
4PF4
 
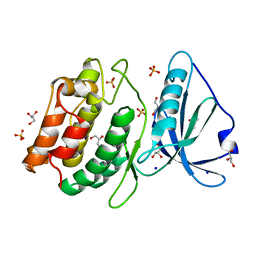 | | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277 | | Descriptor: | Death-associated protein kinase 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Temmerman, K, Simon, B, Wilmanns, M. | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277
To Be Published
|
|
4TL0
 
 | |
1O17
 
 | |
1JQQ
 
 | | Crystal structure of Pex13p(301-386) SH3 domain | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20 | | Authors: | Douangamath, A, Mayans, O, Barnett, P, Distel, B, Wilmanns, M. | | Deposit date: | 2001-08-08 | | Release date: | 2002-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
Mol.Cell, 10, 2002
|
|
6FX5
 
 | | MITF dimerization mutant | | Descriptor: | Microphthalmia-associated transcription factor, SULFATE ION | | Authors: | Pogenberg, V, Milewski, M, Wilmanns, M. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of conditional partner selectivity in MITF/TFE family transcription factors with a conserved coiled coil stammer motif.
Nucleic Acids Res., 48, 2020
|
|
2XIV
 
 | |
2Y4P
 
 | |
2Y4V
 
 | |
2XUU
 
 | | Crystal structure of a DAP-kinase 1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEATH-ASSOCIATED PROTEIN KINASE 1, MAGNESIUM ION, ... | | Authors: | de Diego, I, Kuper, J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
2XQ1
 
 | | Crystal structure of peroxisomal catalase from the yeast Hansenula polymorpha | | Descriptor: | PEROXISOMAL CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Penya-Soler, E, Vega, M.C, Wilmanns, M, Williams, C.P. | | Deposit date: | 2010-08-31 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Features of Peroxisomal Catalase from the Yeast Hansenula Polymorpha
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6G1L
 
 | | MITF/CLEARbox structure | | Descriptor: | CLEAR-box, Microphthalmia-associated transcription factor | | Authors: | Pogenberg, V, Wilmanns, M. | | Deposit date: | 2018-03-21 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MITF has a central role in regulating starvation-induced autophagy in melanoma.
Sci Rep, 9, 2019
|
|
6Y1L
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47R mutant HEAVY CHAIN, FAB A.17 L47R mutant Light CHAIN, ... | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y49
 
 | | Crystal structure of the paraoxon-modified A.17kappa antibody FAB fragment | | Descriptor: | A.17kappa antibody FAB fragment - Heavy Chain, A.17kappa antibody FAB fragment - Light Chain, DIETHYL PHOSPHONATE | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y1N
 
 | | Crystal structure of the phosphonate-modified A.5 antibody FAB fragment | | Descriptor: | 8-METHYL-8-AZABICYCLO[3.2.1]OCTAN-3-YL PHENYLPHOSPHONATE, FAB A.5 Heavy chain, FAB A.5 Light Chain | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y1K
 
 | | Crystal structure of the unmodified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | FAB A.17 L47R mutant Heavy Chain, FAB A.17 L47R mutant Light Chain | | Authors: | Chatziefthimiou, S, Stepanova, A, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
