7D1A
 
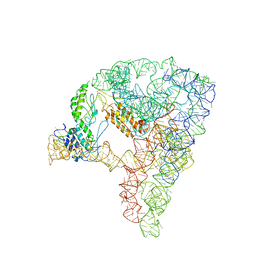 | | cryo-EM structure of a group II intron RNP complexed with its reverse transcriptase | | Descriptor: | Group II intron-encoded protein LtrA, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3'), RNA (692-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
8XRP
 
 | | The Cryo-EM structure of IL-12, receptor subunit beta-1 and receptor subunit beta-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, Interleukin-12 receptor subunit beta-2, ... | | Authors: | Chen, H.Q, Wang, X.Q, Ge, X.F, Zeng, J.W, Wang, J.W. | | Deposit date: | 2024-01-07 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure and assembly of the human IL-12 signaling complex.
Structure, 32, 2024
|
|
3PZ8
 
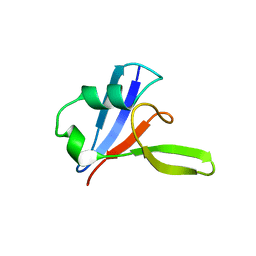 | |
4DDP
 
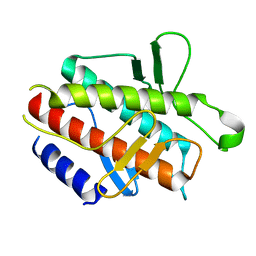 | |
6IIN
 
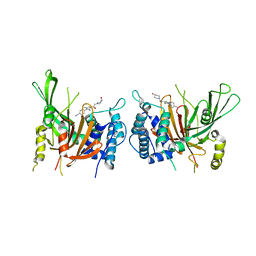 | | USP14 catalytic domain with IU1-248 | | Descriptor: | 4-{3-[(4-hydroxypiperidin-1-yl)acetyl]-2,5-dimethyl-1H-pyrrol-1-yl}benzonitrile, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6IIM
 
 | | USP14 catalytic domain with IU1-206 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(4-hydroxypiperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
8XCG
 
 | | Tail tip complex of bacteriophage lambda in the open state | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Ge, X.F, Wang, J.W. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mechanism of bacteriophage lambda tail's interaction with the bacterial receptor.
Nat Commun, 15, 2024
|
|
8XCI
 
 | |
8XCK
 
 | | Closed state of central tail fiber of bacteriophage lambda | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, Tip attachment protein J | | Authors: | Ge, X.F, Wang, J.W. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural mechanism of bacteriophage lambda tail's interaction with the bacterial receptor.
Nat Commun, 15, 2024
|
|
8XCJ
 
 | |
4M9S
 
 | | crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
4M9X
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.344 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
4M9Y
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
4M9Z
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
8HYJ
 
 | |
3PXG
 
 | | Structure of MecA121 and ClpC1-485 complex | | Descriptor: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.654 Å) | | Cite: | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
7EU0
 
 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 backtracked complex | | Descriptor: | DNA (33-MER), DNA (5'-D(*CP*TP*GP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*GP*AP*GP*TP*G)-3'), DNA-directed RNA polymerase IV subunit 1, ... | | Authors: | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
7EU1
 
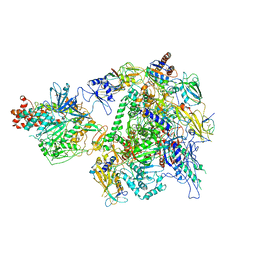 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 holoenzyme | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerases II and IV subunit 5A, DNA-directed RNA polymerases II, ... | | Authors: | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
4LGI
 
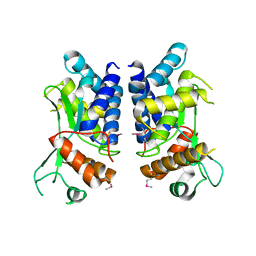 | | N-terminal truncated NleC structure | | Descriptor: | Uncharacterized protein | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7C7R
 
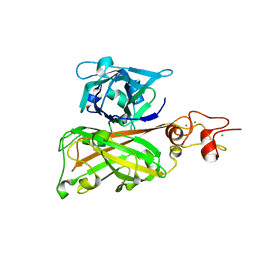 | | Biofilm associated protein - B domain | | Descriptor: | Biofilm-associated surface protein, CALCIUM ION | | Authors: | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
7C7U
 
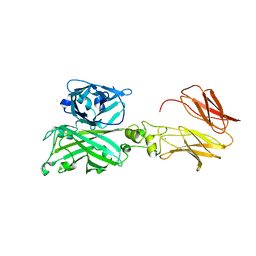 | | Biofilm associated protein - BSP domain | | Descriptor: | Biofilm-associated surface protein, CALCIUM ION | | Authors: | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
