7L8F
 
 | | BG505 SOSIP.v5.2(7S) in complex with the polyclonal Fab pAbC-2 from animal Rh.33172 (Wk38 time point) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Antanasijevic, A, Sewall, L.M, Ward, A.B. | | Deposit date: | 2020-12-31 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM.
Nat Commun, 12, 2021
|
|
1L6G
 
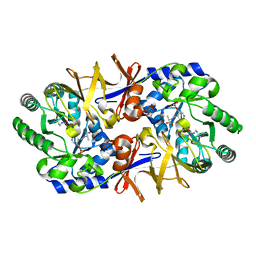 | | Alanine racemase bound with N-(5'-phosphopyridoxyl)-D-alanine | | Descriptor: | N-(5'-PHOSPHOPYRIDOXYL)-D-ALANINE, alanine racemase | | Authors: | Watanabe, A, Yoshimura, T, Mikami, B, Hayashi, H, Kagamiyama, H, Esaki, N. | | Deposit date: | 2002-03-10 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5'-phosphopyridoxyl)alanine.
J.Biol.Chem., 277, 2002
|
|
5B5E
 
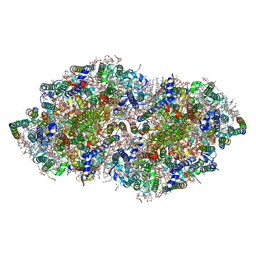 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, A, Fukushima, Y, Kamiya, N. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Two Different Structures of the Oxygen-Evolving Complex in the Same Polypeptide Frameworks of Photosystem II
J. Am. Chem. Soc., 139, 2017
|
|
1L6F
 
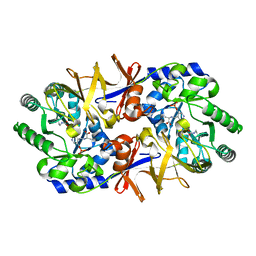 | | Alanine racemase bound with N-(5'-phosphopyridoxyl)-L-alanine | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | Authors: | Watanabe, A, Yoshimura, T, Mikami, B, Hayashi, H, Kagamiyama, H, Esaki, N. | | Deposit date: | 2002-03-09 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5'-phosphopyridoxyl)alanine.
J.Biol.Chem., 277, 2002
|
|
7TQU
 
 | |
7TQT
 
 | |
7TQS
 
 | |
2VSQ
 
 | | Structure of surfactin A synthetase C (SrfA-C), a nonribosomal peptide synthetase termination module | | Descriptor: | LEUCINE, SULFATE ION, SURFACTIN SYNTHETASE SUBUNIT 3 | | Authors: | Tanovic, A, Samel, S.A, Essen, L.-O, Marahiel, M.A. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the termination module of a nonribosomal peptide synthetase.
Science, 321, 2008
|
|
4RE2
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
4RE3
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
4RE4
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Beta-mannosidase/beta-glucosidase, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
8EQN
 
 | |
7SGF
 
 | | Lassa virus glycoprotein construct (Josiah GPC-I53-50A) in complex with LAVA01 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Antanasijevic, A, Brouwer, P.J.M, Ward, A.B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-12 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection.
Cell Host Microbe, 30, 2022
|
|
7SGE
 
 | |
7SGD
 
 | | Lassa virus glycoprotein construct(Josiah GPCysR4) recovered from GPC-I53-50 nanoparticle by localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Antanasijevic, A, Brouwer, P.J.M, Ward, A.B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-12 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection.
Cell Host Microbe, 30, 2022
|
|
8P6U
 
 | | Human carbonic anhydrase II containing 5-fluorotryptophanes | | Descriptor: | BENZOIC ACID, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
8PHL
 
 | | Human carbonic anhydrase II containing 4-fluorophenylalanine | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-06-20 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
8Q0C
 
 | | Human carbonic anhydrase II containing 3-fluorotyrosine | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-07-28 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
6VFJ
 
 | |
6VFH
 
 | |
6VFI
 
 | |
1Z6X
 
 | | Structure Of Human ADP-Ribosylation Factor 4 | | Descriptor: | ADP-ribosylation factor 4, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure Of Human ADP-Ribosylation Factor 4
To be Published
|
|
1ZD9
 
 | | Structure of human ADP-ribosylation factor-like 10B | | Descriptor: | ADP-ribosylation factor-like 10B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 10B
to be published
|
|
1YZG
 
 | | Structure of Human ADP-ribosylation factor-like 8 | | Descriptor: | ADP-ribosylation factor-like 8, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human ADP-ribosylation factor-like 8
To be Published, 2005
|
|
1Z6Y
 
 | | Structure Of Human ADP-Ribosylation Factor-Like 5 | | Descriptor: | ADP-ribosylation factor-like protein 5, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure Of Human ADP-Ribosylation Factor-Like 5
To be Published
|
|
