6DL7
 
 | | Human mitochondrial ClpP in complex with ONC201 (TIC10) | | Descriptor: | 7-benzyl-4-[(2-methylphenyl)methyl]-6,7,8,9-tetrahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(4H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Halgas, O, Zarabi, S.F, Schimmer, A, Pai, E.F. | | Deposit date: | 2018-05-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality.
Cancer Cell, 35, 2019
|
|
6LXU
 
 | |
7N82
 
 | | NMR Solution structure of Se0862 | | Descriptor: | Biofilm-related protein | | Authors: | Zhang, N, LiWang, A.L. | | Deposit date: | 2021-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2.
Proteins, 89, 2021
|
|
3OGN
 
 | |
1GIB
 
 | | MU-CONOTOXIN GIIIB, NMR | | Descriptor: | MU-CONOTOXIN GIIIB | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels.
Biochemistry, 35, 1996
|
|
1HAK
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN PLACENTAL ANNEXIN V COMPLEXED WITH K-201 AS A CALCIUM CHANNEL ACTIVITY INHIBITOR | | Descriptor: | 4-[3-{1-(4-BENZYL)PIPERODINYL}PROPIONYL]-7-METHOXY-2,3,4,5-TERTRAHYDRO-1,4-BENZOTHIAZEPINE, ANNEXIN V | | Authors: | Ago, H, Inagaki, E, Miyano, M. | | Deposit date: | 1997-12-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of annexin V with its ligand K-201 as a calcium channel activity inhibitor.
J.Mol.Biol., 274, 1997
|
|
6MRR
 
 | | De novo designed protein Foldit1 | | Descriptor: | Foldit1 | | Authors: | Koepnick, B, Bick, M.J, Estep, R.D, Kleinfelter, S, Wei, L, Baker, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MRS
 
 | |
6NUK
 
 | | De novo designed protein Ferredog-Diesel | | Descriptor: | Ferredog-Diesel | | Authors: | Koepnick, B, Bick, M.J, DiMaio, F, Norgard-Solano, T, Baker, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
7WKI
 
 | |
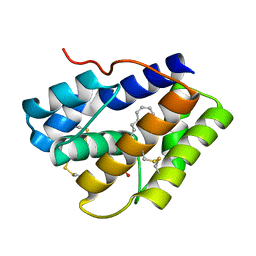
4INW
 
 | |
4INX
 
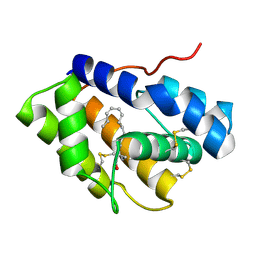 | | Structure of Pheromone-binding protein 1 in complex with (Z,Z)-11,13- hexadecadienol | | Descriptor: | (11Z,13Z)-hexadeca-11,13-dien-1-ol, Pheromone-binding protein 1 | | Authors: | di Luccio, E, Wilson, D.K. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Crystallographic Observation of pH-Induced Conformational Changes in the Amyelois transitella Pheromone-Binding Protein AtraPBP1.
Plos One, 8, 2013
|
|
1XFR
 
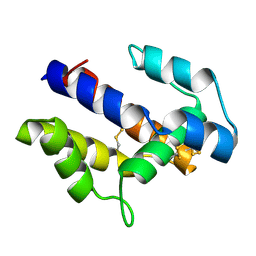 | |
3I7V
 
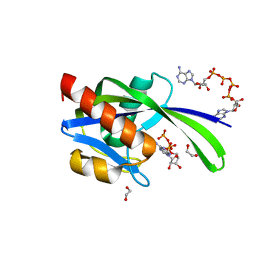 | | Crystal structure of AP4A hydrolase complexed with AP4A (ATP) (aq_158) from Aquifex aeolicus Vf5 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3I7U
 
 | | Crystal structure of AP4A hydrolase (aq_158) from Aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2KPH
 
 | | NMR Structure of AtraPBP1 at pH 4.5 | | Descriptor: | Pheromone binding protein | | Authors: | Ames, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Navel Orangeworm Moth Pheromone-Binding Protein (AtraPBP1): Implications for pH-Sensitive Pheromone Detection .
Biochemistry, 49, 2010
|
|
2ZQE
 
 | | Crystal structure of the Smr domain of Thermus thermophilus MutS2 | | Descriptor: | MutS2 protein | | Authors: | Fukui, K, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2008-08-08 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MutS2 endonuclease domain and the mechanism of homologous recombination suppression
J.Biol.Chem., 283, 2008
|
|
7FG2
 
 | |
7FG3
 
 | |
7FG7
 
 | |
