6ZHS
 
 | | Uba1 bound to two E2 (Ubc13) molecules | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
6ZHU
 
 | | Yeast Uba1 in complex with Ubc3 and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system
To Be Published
|
|
7PVN
 
 | | Crystal Structure of Human UBA6 in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Truongvan, N, Li, S, Schindelin, H. | | Deposit date: | 2021-10-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
7POE
 
 | |
7PUX
 
 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
1FMA
 
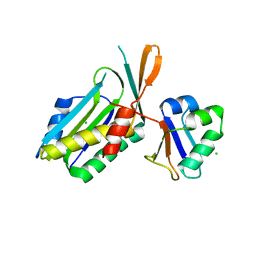 | | MOLYBDOPTERIN SYNTHASE (MOAD/MOAE) | | Descriptor: | CHLORIDE ION, MOLYBDOPTERIN CONVERTING FACTOR, SUBUNIT 1, ... | | Authors: | Rudolph, M.J, Wuebbens, M.M, Rajagolpalan, K.V, Schindelin, H. | | Deposit date: | 2000-08-16 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation.
Nat.Struct.Biol., 8, 2001
|
|
1FR9
 
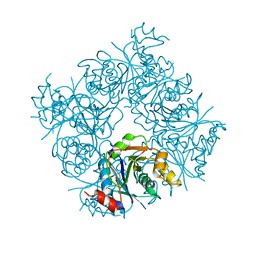 | | STRUCTURE OF E. COLI MOBA | | Descriptor: | ACETATE ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN, ZINC ION | | Authors: | Lake, M.W, Temple, C.A, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-09-07 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the Escherichia coli MobA protein provides insight into molybdopterin guanine dinucleotide biosynthesis.
J.Biol.Chem., 275, 2000
|
|
1FRW
 
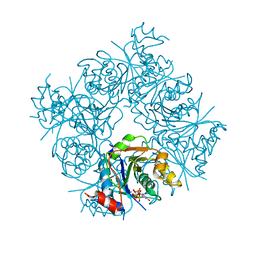 | | STRUCTURE OF E. COLI MOBA WITH BOUND GTP AND MANGANESE | | Descriptor: | ACETATE ION, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Lake, M.W, Temple, C.A, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-09-07 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the Escherichia coli MobA protein provides insight into molybdopterin guanine dinucleotide biosynthesis.
J.Biol.Chem., 275, 2000
|
|
1G8L
 
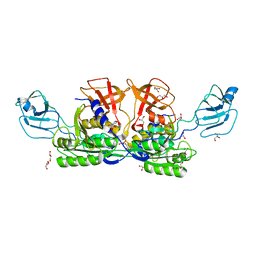 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI MOEA | | Descriptor: | GLYCEROL, MOLYBDOPTERIN BIOSYNTHESIS MOEA PROTEIN | | Authors: | Xiang, S, Nichols, J, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of Escherichia coli MoeA and its relationship to the multifunctional protein gephyrin.
Structure, 9, 2001
|
|
1G8R
 
 | | MOEA | | Descriptor: | GLYCEROL, MOLYBDOPTERIN BIOSYNTHESIS MOEA PROTEIN | | Authors: | Xiang, S, Nichols, J, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-11-20 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of Escherichia coli MoeA and its relationship to the multifunctional protein gephyrin.
Structure, 9, 2001
|
|
6ENS
 
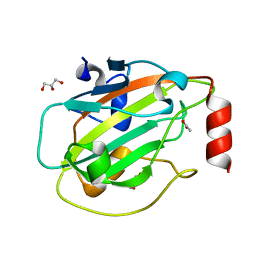 | | Structure of mouse wild-type RKIP | | Descriptor: | ACETATE ION, GLYCEROL, Phosphatidylethanolamine-binding protein 1 | | Authors: | Hirschbeck, M, Koelmel, W, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6FGC
 
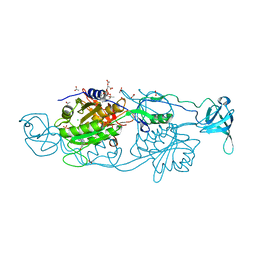 | | Crystal structure of Gephyrin E domain in complex with Artesunate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Kasaragod, V.B, Schindelin, H. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Neuron, 101, 2019
|
|
6GXY
 
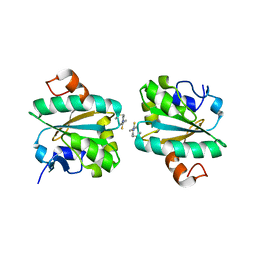 | | Tryparedoxin from Trypanosoma brucei in complex with CFT | | Descriptor: | 5-(4-fluorophenyl)-2-methyl-3~{H}-thieno[2,3-d]pyrimidin-4-one, Tryparedoxin | | Authors: | Bader, N, Wagner, A, Hellmich, U, Schindelin, H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor-Induced Dimerization of an Essential Oxidoreductase from African Trypanosomes.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6FGD
 
 | |
6GXG
 
 | | Tryparedoxin from Trypanosoma brucei in complex with CFT | | Descriptor: | 5-(4-fluorophenyl)-2-methyl-3~{H}-thieno[2,3-d]pyrimidin-4-one, GLYCEROL, Tryparedoxin | | Authors: | Bader, N, Wagner, A, Hellmich, U, Schindelin, H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitor-Induced Dimerization of an Essential Oxidoreductase from African Trypanosomes.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
3BOA
 
 | |
3QQ8
 
 | |
3R3M
 
 | |
3QQ7
 
 | |
4BKM
 
 | | Crystal structure of the murine AUM (phosphoglycolate phosphatase) capping domain as a fusion protein with the catalytic core domain of murine chronophin (pyridoxal phosphate phosphatase) | | Descriptor: | MAGNESIUM ION, NITRATE ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Seifried, A, Gohla, A, Schindelin, H. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evolutionary and Structural Analyses of the Mammalian Haloacid Dehalogenase-Type Phosphatases Aum and Chronophin Provide Insight Into the Basis of Their Different Substrate Specificities.
J.Biol.Chem., 289, 2014
|
|
4BX0
 
 | | Crystal Structure of a Monomeric Variant of murine Chronophin (Pyridoxal Phosphate phosphatase) | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYRIDOXAL PHOSPHATE PHOSPHATASE | | Authors: | Kestler, C, Knobloch, G, Gohla, A, Schindelin, H. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Chronophin Dimerization is Required for Proper Positioning of its Substrate Specificity Loop
J.Biol.Chem., 289, 2014
|
|
4BX3
 
 | |
4BX2
 
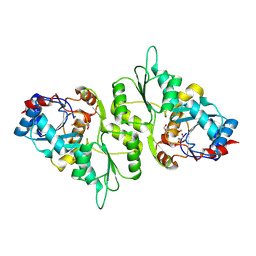 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in complex with Beryllium trifluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Knobloch, G, Gohla, A, Schindelin, H. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Chronophin Dimerization is Required for Proper Positioning of its Substrate Specificity Loop.
J.Biol.Chem., 289, 2014
|
|
4PD1
 
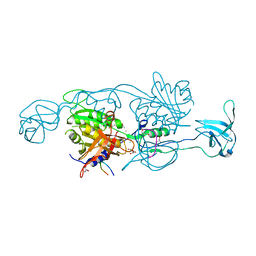 | | Structure of gephyrin E domain with Glycine-beta receptor peptide | | Descriptor: | ACETATE ION, GLYCEROL, Gephyrin, ... | | Authors: | Kasaragod, V.B, Maric, H.M, Schindelin, H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Modulation of gephyrin-glycine receptor affinity by multivalency.
Acs Chem.Biol., 9, 2014
|
|
4OYU
 
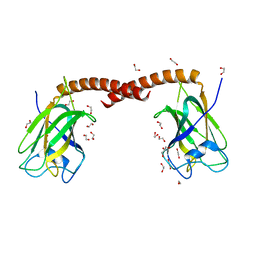 | | Crystal structure of the N-terminal domains of muskelin | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Muskelin | | Authors: | Delto, C, Kuper, J, Schindelin, H. | | Deposit date: | 2014-02-13 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The LisH Motif of Muskelin Is Crucial for Oligomerization and Governs Intracellular Localization.
Structure, 23, 2015
|
|
