4C8S
 
 | |
4C95
 
 | |
4C93
 
 | |
2VCP
 
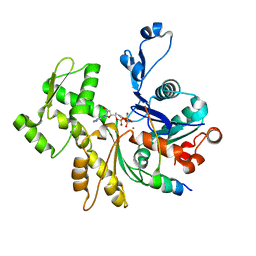 | | Crystal structure of N-Wasp VC domain in complex with skeletal actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gaucher, J.F, Didry, D, Carlier, M.F. | | Deposit date: | 2007-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interactions of isolated C-terminal fragments of neural Wiskott-Aldrich syndrome protein (N-WASP) with actin and Arp2/3 complex.
J. Biol. Chem., 287, 2012
|
|
5HOI
 
 | |
5HOG
 
 | |
5NXQ
 
 | |
5MEC
 
 | |
5MEA
 
 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 2. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5MEB
 
 | | Crystal structure of yeast Cdt1 C-terminal domain | | Descriptor: | Cell division cycle protein CDT1, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5ME9
 
 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 1. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
1K5G
 
 | | Crystal structure of Ran-GDP-AlFx-RanBP1-RanGAP complex | | Descriptor: | ALUMINUM FLUORIDE, GTP-binding nuclear protein RAN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
1K5D
 
 | | Crystal structure of Ran-GPPNHP-RanBP1-RanGAP complex | | Descriptor: | GTP-binding nuclear protein RAN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
