3SLB
 
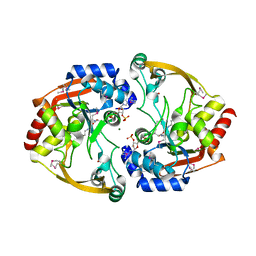 | | Crystal structure of BA2930 in complex with AcCoA and cytosine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of BA2930 in complex with AcCoA and cytosine
TO BE PUBLISHED
|
|
3SLF
 
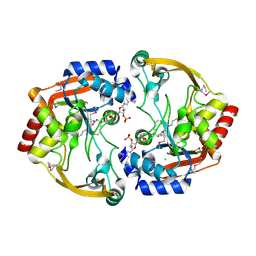 | | Crystal structure of BA2930 in complex with AcCoA and uracil | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of BA2930 in complex with AcCoA and uracil
TO BE PUBLISHED
|
|
3UEH
 
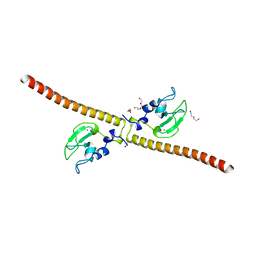 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3V09
 
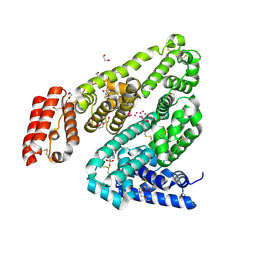 | | Crystal structure of Rabbit Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3UED
 
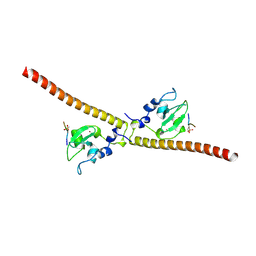 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3 (C2 space group). | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEF
 
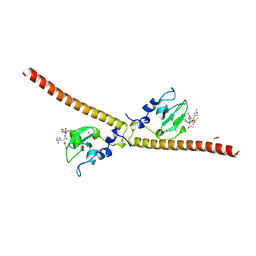 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEE
 
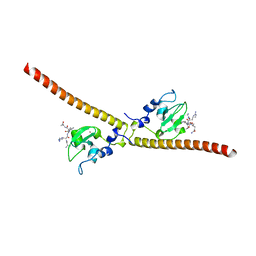 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3V08
 
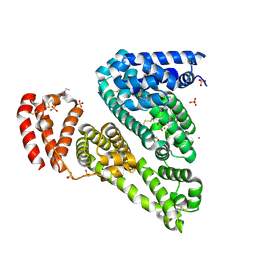 | | Crystal structure of Equine Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, SULFATE ION, ... | | Authors: | Dayal, A, Jablonska, K, Porebski, P.J, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3TPF
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Grimshaw, S, Porebski, P.J, Grabowski, M, Savchenko, A, Chruszcz, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni at 2.7 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEC
 
 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Baculoviral IAP repeat-containing protein 5, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Cooper, D.R, Chruszcz, M, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEI
 
 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3V03
 
 | | Crystal structure of Bovine Serum Albumin | | Descriptor: | ACETATE ION, CALCIUM ION, Serum albumin | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
2QMO
 
 | | Crystal structure of dethiobiotin synthetase (bioD) from Helicobacter pylori | | Descriptor: | CHLORIDE ION, Dethiobiotin synthetase | | Authors: | Chruszcz, M, Xu, X, Cuff, M, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-16 | | Release date: | 2007-07-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
6S0V
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, neamine and sulfate | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0W
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and kanamycin B sulfate | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6MDQ
 
 | | Crystal structure of equine serum albumin in complex with testosterone | | Descriptor: | CITRATE ANION, Serum albumin, TESTOSTERONE | | Authors: | Czub, M.P, Majorek, K.A, Shabalin, I.G, Handing, K.B, Venkataramany, B.S, Cymborowski, M.T, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-05 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Testosterone meets albumin - the molecular mechanism of sex hormone transport by serum albumins.
Chem Sci, 10, 2019
|
|
3IJW
 
 | | Crystal structure of BA2930 in complex with CoA | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3E4F
 
 | | Crystal structure of BA2930- a putative aminoglycoside N3-acetyltransferase from Bacillus anthracis | | Descriptor: | Aminoglycoside N3-acetyltransferase, CITRIC ACID | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
6EDV
 
 | | Structure of a GNAT superfamily acetyltransferase PA3944 in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, CALCIUM ION, ... | | Authors: | Majorek, K.A, Satchell, K.J.F, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-12 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
