1SIO
 
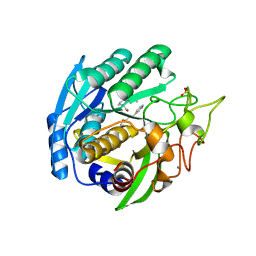 | | Structure of Kumamolisin-As complexed with a covalently-bound inhibitor, AcIPF | | Descriptor: | Ace-ILE-PRO-PHL peptide inhibitor, CALCIUM ION, SULFATE ION, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
1SN7
 
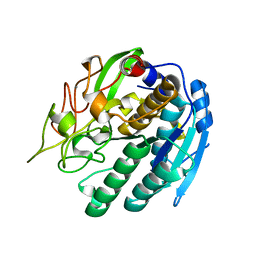 | | KUMAMOLISIN-AS, APOENZYME | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Oda, K, Nishino, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-as, a serine-carboxyl peptidase with collagenase activity.
J.Biol.Chem., 279, 2004
|
|
1SIU
 
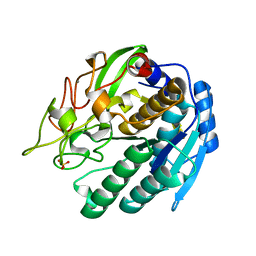 | | KUMAMOLISIN-AS E78H MUTANT | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
1T1G
 
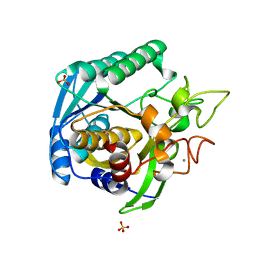 | | High Resolution Crystal Structure of Mutant E23A of Kumamolisin, a sedolisin type proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1T1E
 
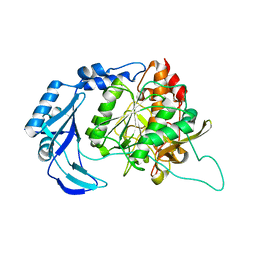 | | High Resolution Crystal Structure of the Intact Pro-Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1T1I
 
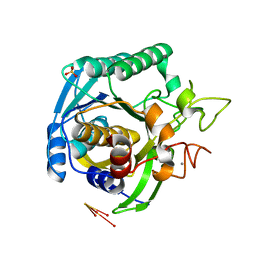 | | High Resolution Crystal Structure of Mutant W129A of Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
8YEJ
 
 | | Cryo-EM structure of the channelrhodopsin GtCCR2 focused on the monomer | | Descriptor: | GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 84, 2024
|
|
8YEK
 
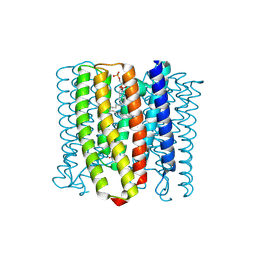 | | Cryo-EM structure of the channelrhodopsin GtCCR2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 84, 2024
|
|
8YEL
 
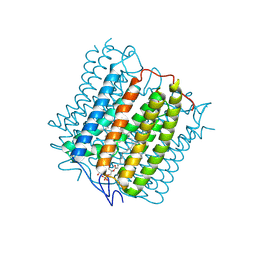 | | Cryo-EM structure of the channelrhodopsin GtCCR4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cation channel rhodopsin 4, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 84, 2024
|
|
1F53
 
 | | NMR STRUCTURE OF KILLER TOXIN-LIKE PROTEIN SKLP | | Descriptor: | YEAST KILLER TOXIN-LIKE PROTEIN | | Authors: | Ohki, S, Kariya, E, Hiraga, K, Wakamiya, A, Isobe, T, Oda, K, Kainosho, M. | | Deposit date: | 2000-06-12 | | Release date: | 2000-12-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Streptomyces killer toxin-like protein, SKLP: further evidence for the wide distribution of single-domain betagamma-crystallin superfamily proteins.
J.Mol.Biol., 305, 2001
|
|
3X44
 
 | | Crystal structure of O-ureido-L-serine-bound K43A mutant of O-ureido-L-serine synthase | | Descriptor: | (E)-O-(carbamoylamino)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, O-ureido-L-serine synthase | | Authors: | Matoba, Y, Uda, N, Oda, K, Sugiyama, M. | | Deposit date: | 2015-03-13 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural and mutational analyses of O-ureido-L-serine synthase necessary for D-cycloserine biosynthesis.
Febs J., 282, 2015
|
|
3X43
 
 | | Crystal structure of O-ureido-L-serine synthase | | Descriptor: | O-ureido-L-serine synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matoba, Y, Uda, N, Oda, K, Sugiyama, M. | | Deposit date: | 2015-03-13 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structural and mutational analyses of O-ureido-L-serine synthase necessary for D-cycloserine biosynthesis.
Febs J., 282, 2015
|
|
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GTL
 
 | | The thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Pro-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GTJ
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GT9
 
 | | High resolution crystal structure of a thermostable serine-carboxyl type proteinase, kumamolisin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN, SULFATE ION | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-14 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1S2K
 
 | | Structure of SCP-B a member of the Eqolisin family of Peptidases in a complex with a Tripeptide Ala-Ile-His | | Descriptor: | Ala-Ile-His tripeptide, Scytalidopepsin B, TYROSINE | | Authors: | Fujinaga, M, Cherney, M.M, Oyama, H, Oda, K, James, M.N. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S2B
 
 | | Structure of SCP-B the first member of the Eqolisin family of Peptidases to have its structure determined | | Descriptor: | Scytalidopepsin B | | Authors: | Fujinaga, M, Cherney, M.M, Oyama, H, Oda, K, James, M.N. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6JMQ
 
 | | LAT1-CD98hc complex bound to MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Nakane, T, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JMR
 
 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6K54
 
 | | Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with trisaccharide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Novel hyperthermophilic cellobiohydrolase II isolated from hot spring microbial community
To Be Published
|
|
6K53
 
 | | A variant of metagenome-derived GH6 cellobiohydrolase, HmCel6A (P88S/L230F/F414S) | | Descriptor: | CITRATE ANION, GH6 cellobiohydrolase, HMCEL6A, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from a hot spring metagenomic data
To Be Published
|
|
6K55
 
 | | Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide | | Descriptor: | Glucanase, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
6K52
 
 | | Hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from the microbial flora of a Japanese hot spring | | Descriptor: | ACETATE ION, CALCIUM ION, GH6 cellobiohydrolase, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68000138 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
8I3Y
 
 | | Crystal structure of ASCT from Trypanosoma brucei in complex with Succinyl-CoA. | | Descriptor: | CALCIUM ION, SUCCINIC ACID, SUCCINYL-COENZYME A, ... | | Authors: | Mochizuki, K, Inaoka, D.K, Fukuda, K, Kurasawa, H, Iyoda, K, Nakai, U, Harada, S, Balogun, E.O, Mazet, M, Millerioux, Y, Bringaud, F, Boshart, M, Hirayama, K, Kita, K, Shiba, T. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ASCT from Trypanosoma brucei in complex with Succinyl-CoA.
To Be Published
|
|
