1Z63
 
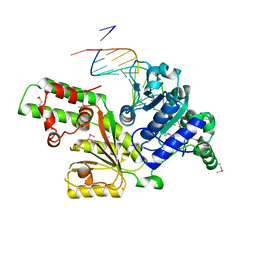 | | Sulfolobus solfataricus SWI2/SNF2 ATPase core in complex with dsDNA | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*A*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*AP*AP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*TP*TP*TP*TP*TP*T)-3', Helicase of the snf2/rad54 family | | Authors: | Duerr, H, Koerner, C, Mueller, M, Hickmann, V, Hopfner, K.P. | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA.
Cell(Cambridge,Mass.), 121, 2005
|
|
6G45
 
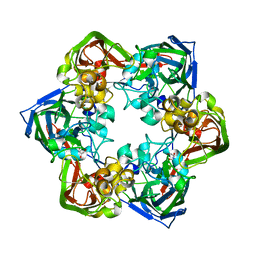 | | Crystal structure of mavirus major capsid protein | | Descriptor: | GLYCEROL, Putative major capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G41
 
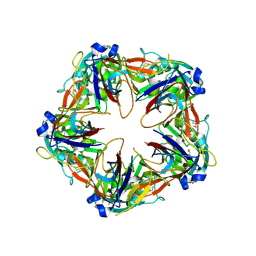 | | Crystal structure of SeMet-labeled mavirus penton protein | | Descriptor: | Minor capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G42
 
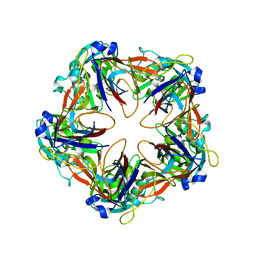 | | Crystal structure of mavirus penton protein | | Descriptor: | Minor capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4A7H
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 2) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
6G43
 
 | | Crystal structure of SeMet-labeled mavirus major capsid protein lacking the C-terminal domain | | Descriptor: | Putative major capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G44
 
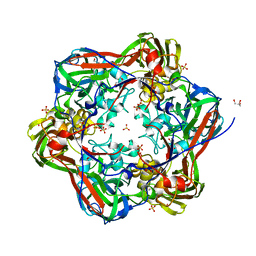 | | Crystal structure of mavirus major capsid protein lacking the C-terminal domain | | Descriptor: | GLYCEROL, Putative major capsid protein, SULFATE ION | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4A7F
 
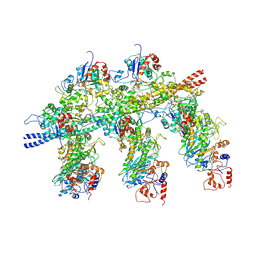 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 3) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
5A3K
 
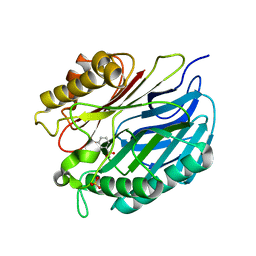 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-HYDROXYBENZOIC ACID, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, SULFATE ION | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
5JLF
 
 | |
4A7N
 
 | | Structure of bare F-actin filaments obtained from the same sample as the Actin-Tropomyosin-Myosin Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, F-ACTIN | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4D5E
 
 | | Crystal Structure of recombinant wildtype CDH | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Loschonsky, S, Wacker, T, Waltzer, S, Giovannini, P.P, McLeish, M.J, Andrade, S.L.A, Mueller, M. | | Deposit date: | 2014-11-03 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Extended Reaction Scope of Thiamine Diphosphate Dependent Cyclohexane-1,2-Dione Hydrolase: From C-C Bond Cleavage to C-C Bond Ligation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4D5G
 
 | | Structure of recombinant CDH-H28AN484A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CYCLOHEXANE-1,2-DIONE HYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loschonsky, S, Wacker, T, Waltzer, S, Giovannini, P.P, McLeish, M.J, Andrade, S.L.A, Mueller, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extended Reaction Scope of Thiamine Diphosphate Dependent Cyclohexane-1,2-Dione Hydrolase: From C-C Bond Cleavage to C-C Bond Ligation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8RPI
 
 | | JanthE from Janthinobacterium sp. HH01, lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | JhE from Janthinobacterium sp. HH01
To Be Published
|
|
7Q03
 
 | | Ketol-acid reductoisomerase from Methanothermococcus thermolithotrophicus in the close state with NADP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, GLYCEROL, ... | | Authors: | Lemaire, O.N, Mueller, M, Wagner, T. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Rearrangements of a Dodecameric Ketol-Acid Reductoisomerase Isolated from a Marine Thermophilic Methanogen.
Biomolecules, 11, 2021
|
|
7Q07
 
 | | Ketol-acid reductoisomerase from Methanothermococcus thermolithotrophicus in the open state with NADP and tartrate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ketol-Acid Reductoisomerase from Methanothermococcus thermolithotrophicus, ... | | Authors: | Lemaire, O.N, Mueller, M, Wagner, T. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Rearrangements of a Dodecameric Ketol-Acid Reductoisomerase Isolated from a Marine Thermophilic Methanogen.
Biomolecules, 11, 2021
|
|
2Y1J
 
 | | CRYSTAL STRUCTURE OF A R-DIASTEREOMER ANALOGUE OF THE SPORE PHOTOPRODUCT IN COMPLEX WITH FRAGMENT DNA POLYMERASE I FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | 5'-D(*AP*GP*GP*GP*QBTP*THM*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*AP*CP*CP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Heil, K, Schneider, S, Mueller, M, Kneuttinger, A.C, Carell, T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the Uv-Induced Spore Photoproduct DNA Lesion.
Chemistry, 17, 2011
|
|
2Y1I
 
 | | Crystal structure of a S-diastereomer analogue of the spore photoproduct in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*AP*GP*GP*GP*PBTP*THM*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*AP*CP*CP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Heil, K, Schneider, S, Mueller, M, Kneuttinger, A.C, Carell, T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the Uv-Induced Spore Photoproduct DNA Lesion.
Chemistry, 17, 2011
|
|
5N91
 
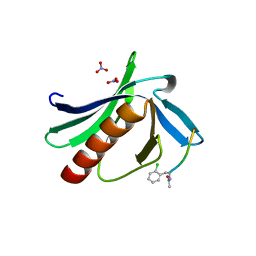 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPP-OH | | Descriptor: | Ac-[2-Cl-F]-PPPP-OH, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NCG
 
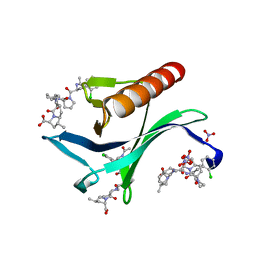 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-9]-OH | | Descriptor: | (3~{S},7~{R},10~{R},11~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-11-methyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-04 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NAJ
 
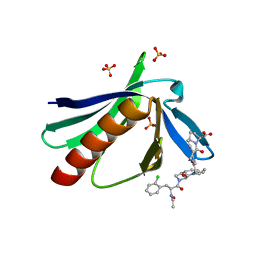 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-1]-[ProM-1]-OH | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{R},7~{S},10~{S},13~{R})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{S},7~{R},10~{R},13~{S})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, CHLORIDE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NBF
 
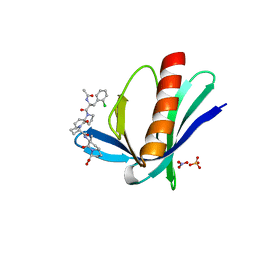 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-3]-OH | | Descriptor: | (3~{S},6~{S},7~{R},9~{a}~{R})-6-[[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonylamino]-7-methyl-5-oxidanylidene-1,2,3,6,7,9~{a}-hexahydropyrrolo[1,2-a]azepine-3-carboxylic acid, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NC7
 
 | | ENAH EVH1 in complex with Ac-WPPPPTEDEL-NH2 | | Descriptor: | ActA-derived 10-mer Ac-FPPPPTEDEL-NH2 with acetylated (Ac) and amidated (NH2) termini. Phe is substitued by Trp to increase affinity for crystallization, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-03 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NEG
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-13]-OEt | | Descriptor: | NITRATE ION, Protein enabled homolog, SULFATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
