4GG2
 
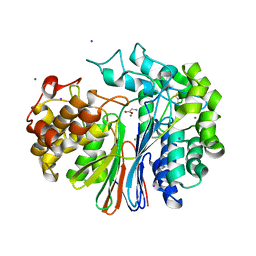 | | The crystal structure of glutamate-bound human gamma-glutamyltranspeptidase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-04 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
4GDX
 
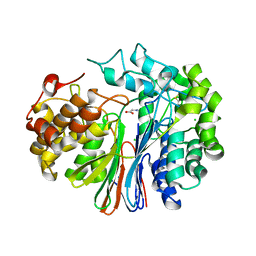 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
1DCV
 
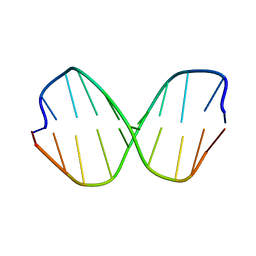 | |
1DCW
 
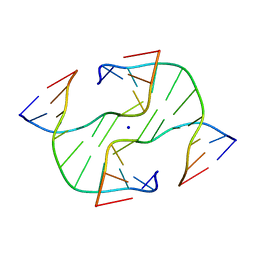 | | STRUCTURE OF A FOUR-WAY JUNCTION IN AN INVERTED REPEAT SEQUENCE. | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3'), SODIUM ION | | Authors: | Eichman, B.F, Vargason, J.M, Mooers, B.H.M, Ho, P.S. | | Deposit date: | 1999-11-05 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1LPY
 
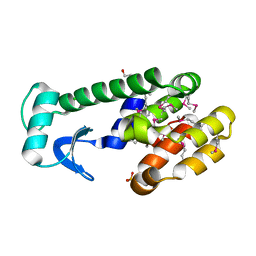 | | Multiple Methionine Substitutions in T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME, ... | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
1LWK
 
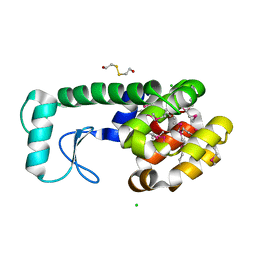 | | Multiple Methionine Substitutions are Tolerated in T4 Lysozyme and have Coupled Effects on Folding and Stability | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.M. | | Deposit date: | 2002-05-31 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
1LWG
 
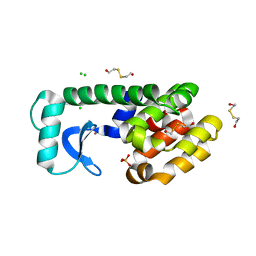 | | Multiple Methionine Substitutions are Tolerated in T4 Lysozyme and have Coupled Effects on Folding and Stability | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.M. | | Deposit date: | 2002-05-31 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
1LW9
 
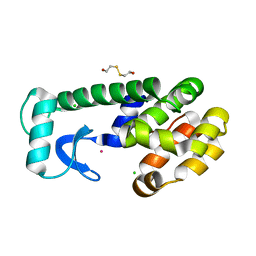 | | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-05-30 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
