7SEM
 
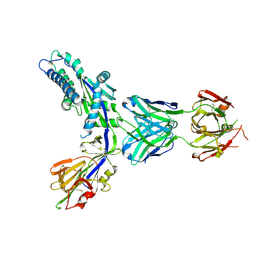 | | Structure-based design of prefusion-stabilized human metapneumovirus fusion proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, MPE8 Fab heavy chain, ... | | Authors: | Rush, S.A, Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of prefusion-stabilized human metapneumovirus fusion proteins.
Nat Commun, 13, 2022
|
|
5VYH
 
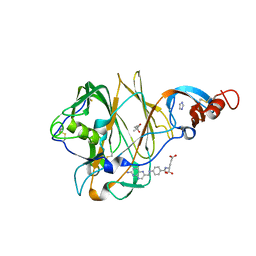 | | Crystal Structure of MERS-CoV S1 N-terminal Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Wang, N, Wrapp, D, Pallesen, J, Ward, A.B, McLellan, J.S. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8VAO
 
 | |
8V5K
 
 | | Structure of the Human Respirovirus 3 Fusion Protein Bound to Camelid Nanobodies 4C03 and 4C06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Camelid Nanobody 4C03, Camelid Nanobody 4C06, ... | | Authors: | Johnson, N.V, Ramamohan, A.R, McLellan, J.S. | | Deposit date: | 2023-11-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for potent neutralization of human respirovirus type 3 by protective single-domain camelid antibodies.
Nat Commun, 15, 2024
|
|
8V5A
 
 | |
8V62
 
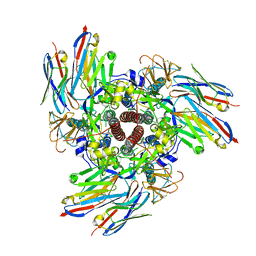 | |
7S3M
 
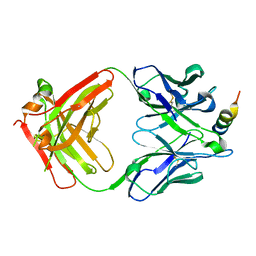 | |
7RW6
 
 | | BORF2-APOBEC3Bctd Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, Maltose/maltodextrin-binding periplasmic protein,Ribonucleoside-diphosphate reductase large subunit, ZINC ION | | Authors: | Shaban, N.M, Yan, R, Shi, K, McLellan, J.S, Yu, Z, Harris, R.S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure of the EBV ribonucleotide reductase BORF2 and mechanism of APOBEC3B inhibition.
Sci Adv, 8, 2022
|
|
6EAH
 
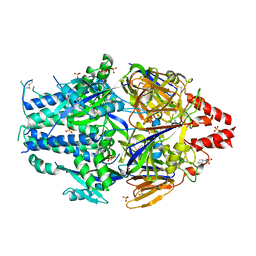 | |
6EAL
 
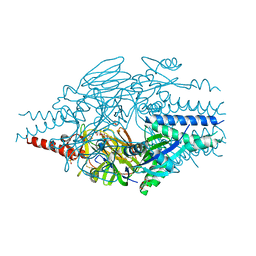 | |
6EAM
 
 | |
6EAD
 
 | |
6EAF
 
 | |
6EAK
 
 | |
6EAE
 
 | |
6EAJ
 
 | |
5WB0
 
 | |
6EAI
 
 | |
6U7K
 
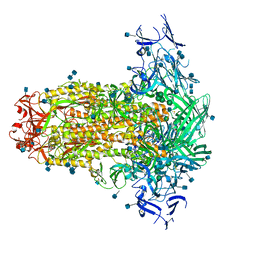 | | Prefusion structure of PEDV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-09-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The 3.1-Angstrom Cryo-electron Microscopy Structure of the Porcine Epidemic Diarrhea Virus Spike Protein in the Prefusion Conformation.
J.Virol., 93, 2019
|
|
3ONY
 
 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with Fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-08-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of GII.10 and GII.12 Norovirus Protruding Domains in Complex with Histo-Blood Group Antigens Reveal Details for a Potential Site of Vulnerability.
J.Virol., 85, 2011
|
|
3ONU
 
 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-08-30 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Crystal Structures of GII.10 and GII.12 Norovirus Protruding Domains in Complex with Histo-Blood Group Antigens Reveal Details for a Potential Site of Vulnerability.
J.Virol., 85, 2011
|
|
3PA1
 
 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with HBGA type A | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, IMIDAZOLE, ... | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-10-18 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structures of GII.10 and GII.12 Norovirus Protruding Domains in Complex with Histo-Blood Group Antigens Reveal Details for a Potential Site of Vulnerability.
J.Virol., 85, 2011
|
|
3Q38
 
 | | Crystal structure of P domain from norwalk virus strain vietnam 026 in complex with HBGA type B (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, IMIDAZOLE, ... | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
3Q3A
 
 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with HBGA type H2 (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, IMIDAZOLE, ... | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
3PA2
 
 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with HBGA type Ley | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, IMIDAZOLE, ... | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-10-18 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structures of GII.10 and GII.12 Norovirus Protruding Domains in Complex with Histo-Blood Group Antigens Reveal Details for a Potential Site of Vulnerability.
J.Virol., 85, 2011
|
|
