1D6N
 
 | | TERNARY COMPLEX STRUCTURE OF HUMAN HGPRTASE, PRPP, MG2+, AND THE INHIBITOR HPP REVEALS THE INVOLVEMENT OF THE FLEXIBLE LOOP IN SUBSTRATE BINDING | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 3H-PYRAZOLO[4,3-D]PYRIMIDIN-7-OL, MAGNESIUM ION, ... | | Authors: | Balendiran, G.K. | | Deposit date: | 1999-10-14 | | Release date: | 1999-12-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ternary complex structure of human HGPRTase, PRPP, Mg2+, and the inhibitor HPP reveals the involvement of the flexible loop in substrate binding.
Protein Sci., 8, 1999
|
|
3M0U
 
 | |
3M0Q
 
 | |
3M0P
 
 | |
3M0S
 
 | |
3M0R
 
 | |
3M0T
 
 | |
3O31
 
 | | E81Q mutant of MtNAS in complex with a reaction intermediate | | Descriptor: | BROMIDE ION, N-[(3S)-3-amino-3-carboxypropyl]-L-glutamic acid, ThermoNicotianamine Synthase | | Authors: | Dreyfus, C, Pignol, D, Arnoux, P. | | Deposit date: | 2010-07-23 | | Release date: | 2011-06-08 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystallographic structure of thermoNicotianamine synthase with a synthetic reaction intermediate highlights the sequential processing mechanism.
Chem.Commun.(Camb.), 47, 2011
|
|
6RW0
 
 | | Crystal structure of ANGEL2, a 2',3'-cyclic phosphatase | | Descriptor: | GLYCEROL, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ANGEL2 is a member of the CCR4 family of deadenylases with 2',3'-cyclic phosphatase activity.
Science, 369, 2020
|
|
6RVZ
 
 | | Crystal structure of ANGEL2, a 2',3'-cyclic phosphatase, in complex with adenosine-2',3'-vanadate | | Descriptor: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ANGEL2 is a member of the CCR4 family of deadenylases with 2',3'-cyclic phosphatase activity.
Science, 369, 2020
|
|
6Z9U
 
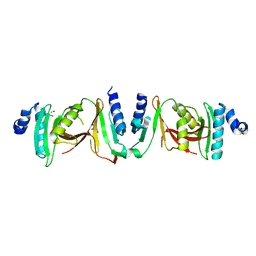 | | Crystal structure of a TSEN15-34 heterodimer. | | Descriptor: | GLYCEROL, tRNA-splicing endonuclease subunit Sen15, tRNA-splicing endonuclease subunit Sen34 | | Authors: | Trowitzsch, S, Sekulovski, S. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.10001779 Å) | | Cite: | Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia.
Nat Commun, 12, 2021
|
|
1E74
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R11E | | Descriptor: | ALPHA-CONOTOXIN IM1(R11E) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E76
 
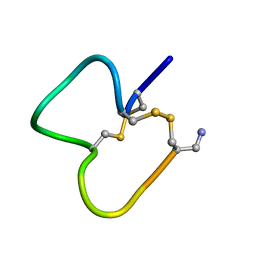 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT D5N | | Descriptor: | ALPHA-CONOTOXIN IM1(D5N) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E75
 
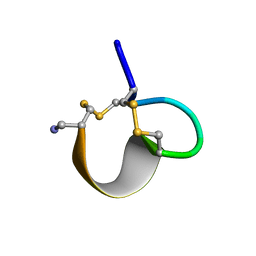 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R7L | | Descriptor: | ALPHA-CONOTOXIN IM1(R7L) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1IM1
 
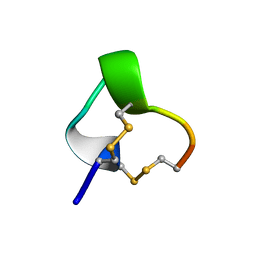 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
