5TMA
 
 | | Zymomonas mobilis pyruvate decarboxylase mutant PDC-2.3 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An iterative computational design approach to increase the thermal endurance of a mesophilic enzyme.
Biotechnol Biofuels, 11, 2018
|
|
6VSP
 
 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase mutant Q247A | | Descriptor: | 1,2-ETHANEDIOL, 2,3-butanediol dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
5UIZ
 
 | | Structure of T.fusca AA10A | | Descriptor: | AA10A, COPPER (II) ION, GLYCEROL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2017-01-16 | | Release date: | 2017-02-01 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Thermobifida fusca lytic polysaccharide monooxygenase and mutagenesis of key residues.
Biotechnol Biofuels, 10, 2017
|
|
5ECU
 
 | |
4WA0
 
 | |
4XEB
 
 | | The structure of P. funicolosum Cel7A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2014-12-23 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering enhanced cellobiohydrolase activity
Nat Commun, 9(1), 2018
|
|
3T9G
 
 | | The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2011-08-02 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A 1.5 A resolution X-ray structure of the catalytic module of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5KX6
 
 | | The structure of Arabidopsis thaliana FUT1 Mutant R284K in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism.
Plant J., 91, 2017
|
|
5KWK
 
 | | The structure of Arabidopsis thaliana FUT1 in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-07-18 | | Release date: | 2016-09-28 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism.
Plant J., 91, 2017
|
|
5KOE
 
 | | The structure of Arabidopsis thaliana FUT1 in complex with XXLG | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism.
Plant J., 91, 2017
|
|
4ITK
 
 | | The structure of C.reinhardtii Ferredoxin 2 | | Descriptor: | Apoferredoxin, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure and biochemical characterization of Chlamydomonas FDX2 reveal two residues that, when mutated, partially confer FDX2 the redox potential and catalytic properties of FDX1.
Photosyn. Res., 128, 2016
|
|
4JJJ
 
 | | The structure of T. fusca GH48 D224N mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2013-03-07 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cel48A from Thermobifida fusca: structure and site directed mutagenesis of key residues.
Biotechnol.Bioeng., 111, 2014
|
|
4LGN
 
 | | The structure of Acidothermus cellulolyticus family 74 glycoside hydrolase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellulose-binding, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Acidothermus cellulolyticus family 74 glycoside hydrolase at 1.82 angstrom resolution.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4DOD
 
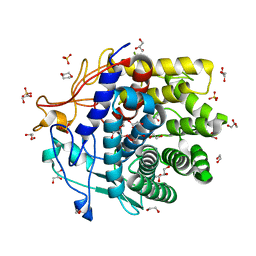 | | The structure of Cbescii CelA GH9 module | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1,4-beta-glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-02-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA.
Science, 342, 2013
|
|
4DOE
 
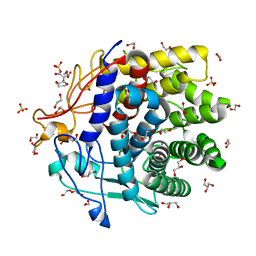 | | The liganded structure of Cbescii CelA GH9 module | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1,4-beta-glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-02-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA.
Science, 342, 2013
|
|
4EL8
 
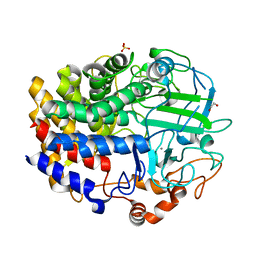 | | The unliganded structure of C.bescii CelA GH48 module | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycoside hydrolase family 48, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-04-10 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA.
Science, 342, 2013
|
|
4EW9
 
 | | The liganded structure of C. bescii family 3 pectate lyase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, ACETATE ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-04-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure and mode of action of Caldicellulosiruptor bescii family 3 pectate lyase in biomass deconstruction.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4FUS
 
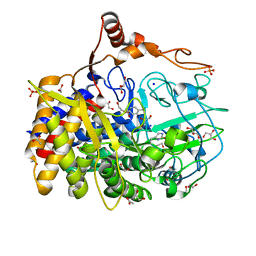 | | The X-ray structure of Hahella chejuensis family 48 glycosyl hydrolase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Sequence, structure, and evolution of cellulases in glycoside hydrolase family 48.
J.Biol.Chem., 287, 2012
|
|
1KK9
 
 | | CRYSTAL STRUCTURE OF E. COLI YCIO | | Descriptor: | SULFATE ION, probable translation factor yciO | | Authors: | Jia, J, Lunin, V.V, Sauve, V, Huang, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-06 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the YciO protein from Escherichia coli
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
6XEX
 
 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase mutant Q247A/V139Q | | Descriptor: | 1,2-ETHANEDIOL, 2,3-butanediol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2020-06-14 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
6XEW
 
 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase | | Descriptor: | 2,3-butanediol dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2020-06-14 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
7M6B
 
 | | The Crystal Structure of Mcbe1 | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2021-03-25 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
3P6B
 
 | | The crystal structure of CelK CBM4 from Clostridium thermocellum | | Descriptor: | ACETATE ION, CALCIUM ION, Cellulose 1,4-beta-cellobiosidase, ... | | Authors: | Alahuhta, P.M, Luo, Y, Lunin, V.V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of CBM4 from Clostridium thermocellum cellulase K.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3PDG
 
 | |
3PE9
 
 | |
