5HOH
 
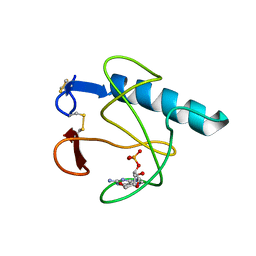 | | RIBONUCLEASE T1 (ASN9ALA/THR93ALA DOUBLEMUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
1LUL
 
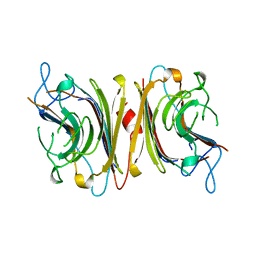 | | DB58, A LEGUME LECTIN FROM DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, LECTIN DB58, MANGANESE (II) ION | | Authors: | Hamelryck, T.W, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M, Loris, R. | | Deposit date: | 1998-06-30 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
3TCJ
 
 | |
3DCQ
 
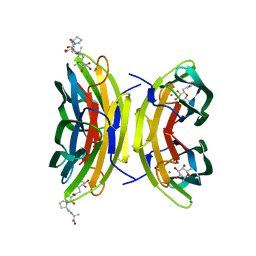 | | LECB (PA-LII) in complex with the synthetic ligand 2G0 | | Descriptor: | (2S)-1-[(2S)-6-amino-2-({[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl]acetyl}amino)hexanoyl]-N-[(1S)-1-carbamoyl-3-methylbutyl]pyrrolidine-2-carboxamide, CALCIUM ION, Fucose-binding lectin PA-IIL | | Authors: | Johansson, E.M, Crusz, S.A, Kolomiets, E, Buts, L, Kadam, R.U, Cacciarini, M, Bartels, K.M, Diggle, S.P, Camara, M, Williams, P, Loris, R, Nativi, C, Rosenau, F, Jaeger, K.E, Darbre, T, Reymond, J.L. | | Deposit date: | 2008-06-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB.
Chem.Biol., 15, 2008
|
|
3DD7
 
 | |
7R5A
 
 | |
7NKV
 
 | |
4QO1
 
 | | p53 DNA binding domain in complex with Nb139 | | Descriptor: | Cellular tumor antigen p53, Nb139 Nanobody against the DNA-binding domain of p53, ZINC ION | | Authors: | De Gieter, S, Bethuyne, J, Gettemans, J, Garcia-Pino, A, Loris, R. | | Deposit date: | 2014-06-19 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | A nanobody modulates the p53 transcriptional program without perturbing its functional architecture.
Nucleic Acids Res., 42, 2014
|
|
4MZP
 
 | | MazF from S. aureus crystal form III, C2221, 2.7 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZM
 
 | | MazF from S. aureus crystal form I, P212121, 2.1 A | | Descriptor: | mRNA interferase MazF | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZT
 
 | | MazF from S. aureus crystal form II, C2221, 2.3 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
6F8S
 
 | | Toxin-Antitoxin complex GraTA | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative Killer protein, SULFATE ION, ... | | Authors: | Talavera, A, Loris, R. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun, 10, 2019
|
|
6F8H
 
 | | antitoxin GraA | | Descriptor: | XRE family transcriptional regulator | | Authors: | Talavera, A, Loris, R. | | Deposit date: | 2017-12-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun, 10, 2019
|
|
6FIX
 
 | | antitoxin GraA in complex with its operator | | Descriptor: | DNA (30-MER), XRE family transcriptional regulator | | Authors: | Talavera, A, Loris, R. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun, 10, 2019
|
|
1G7Y
 
 | | THE CRYSTAL STRUCTURE OF THE 58KD VEGETATIVE LECTIN FROM THE TROPICAL LEGUME DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, STEM/LEAF LECTIN DB58, ... | | Authors: | Buts, L, Hamelryck, T.W, Loris, R, Dao-Thi, M.-H, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
8BT1
 
 | | YdaT transcription regulator (CII functional analog) | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Prolic-Kalinsek, M, Loris, R. | | Deposit date: | 2022-11-27 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39788437 Å) | | Cite: | Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8C7K
 
 | |
3ZBE
 
 | |
1LES
 
 | | LENTIL LECTIN COMPLEXED WITH SUCROSE | | Descriptor: | CALCIUM ION, LENTIL LECTIN, MANGANESE (II) ION, ... | | Authors: | Hamelryck, T, Loris, R. | | Deposit date: | 1995-08-23 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NMR, molecular modeling, and crystallographic studies of lentil lectin-sucrose interaction.
J.Biol.Chem., 270, 1995
|
|
2Z70
 
 | | E.coli RNase 1 in complex with d(CGCGATCGCG) | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DTP*DCP*DGP*DCP*DG)-3'), Ribonuclease I | | Authors: | Martinez-Rodriguez, S, Loris, R, Messens, J. | | Deposit date: | 2007-08-09 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonspecific base recognition mediated by water bridges and hydrophobic stacking in ribonuclease I from Escherichia coli
Protein Sci., 17, 2008
|
|
8OJQ
 
 | |
8OJ9
 
 | |
8OJE
 
 | |
8OJF
 
 | |
3JRZ
 
 | | CcdBVfi-FormII-pH5.6 | | Descriptor: | CcdB | | Authors: | De Jonge, N, Buts, L, Loris, R. | | Deposit date: | 2009-09-09 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic characterization of vibrio fischeri CCDB
J.Biol.Chem., 285, 2010
|
|
