7EQK
 
 | | Structural and mechanistic studies of a novel non-heme iron epimerase/lyase and its utilization in chemoselective synthesis. | | Descriptor: | (E)-3-(1H-indol-3-yl)-2-oxidanyl-but-2-enoic acid, 1-(1~{H}-indol-3-yl)ethanone, Cupin domain-containing protein, ... | | Authors: | Li, T.L, Li, Y.S, Chen, M.H. | | Deposit date: | 2021-05-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04001021 Å) | | Cite: | Structural and Mechanistic Bases for StnK3 and Its Mutant-Mediated Lewis-Acid-Dependent Epimerization and Retro-Aldol Reactions.
Acs Catalysis, 12, 2022
|
|
7EUE
 
 | |
7F6X
 
 | | Structural and mechanistic studies of a novel non-heme iron epimerase/lyase and its utilization in chemoselective synthesis. | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, Cupin domain-containing protein, FE (III) ION, ... | | Authors: | Li, T.L, Li, Y.S, Chen, M.H. | | Deposit date: | 2021-06-26 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and Mechanistic Bases for StnK3 and Its Mutant-MediatedLewis-Acid-Dependent Epimerization and Retro-Aldol Reactions.
Acs Catalysis, 12, 2022
|
|
7EUP
 
 | | Structural and mechanistic studies of a novel non-heme iron epimerase/lyase and its utilization in chemoselective synthesis. | | Descriptor: | (2S,3R)-2-azanyl-3-phenyl-butanoic acid, Cupin domain-containing protein, FE (III) ION | | Authors: | Li, T.L, Li, Y.S, Chen, M.H. | | Deposit date: | 2021-05-18 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11043429 Å) | | Cite: | Structural and Mechanistic Bases for StnK3 and Its Mutant-Mediated Lewis-Acid-Dependent Epimerization and Retro-Aldol Reactions.
Acs Catalysis, 12, 2022
|
|
3LOY
 
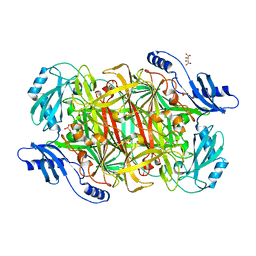 | | Crystal structure of a Copper-containing benzylamine oxidase from Hansenula Polymorpha | | Descriptor: | COPPER (II) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Klema, V.J, Johnson, B.J, Wilmot, C.M. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of substrate specificity in two copper amine oxidases from Hansenula polymorpha.
Biochemistry, 49, 2010
|
|
6L1Y
 
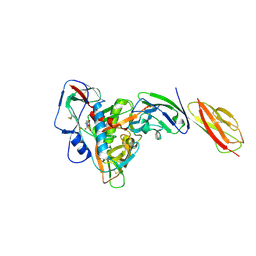 | | structure of gp120/CD4 with a non-canonical surface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, T-cell surface glycoprotein CD4, ... | | Authors: | Liu, X, Ning, W. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | A non-canonical binding interface in the crystal structure of HIV-1 gp120 core in complex with CD4.
Sci Rep, 7, 2017
|
|
8RPO
 
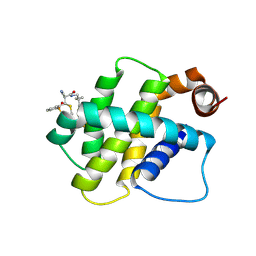 | | BFL1 in complex with a reversible covalent ligand | | Descriptor: | (~{E})-2-cyano-3-(2-dimethylphosphorylphenyl)-~{N}-[[1-[4-(trifluoromethyl)phenyl]cyclopropyl]methyl]prop-2-enamide, Bcl-2-related protein A1 | | Authors: | Hargreaves, D. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Identification and Evaluation of Reversible Covalent Binders to Cys55 of Bfl-1 from a DNA-Encoded Chemical Library Screen.
Acs Med.Chem.Lett., 15, 2024
|
|
8SHJ
 
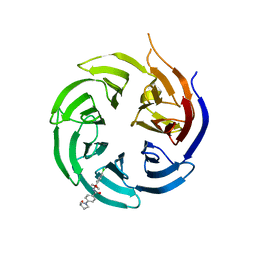 | | Crystal structure of the WD-repeat domain of human WDR91 in complex with MR45279 | | Descriptor: | N-[3-(4-chlorophenyl)oxetan-3-yl]-4-[(3S)-3-hydroxypyrrolidin-1-yl]benzamide, WD repeat-containing protein 91 | | Authors: | Ahmad, H, Zeng, H, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Xu, J, Feng, J.W, Brown, P.J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
7VYY
 
 | | The crystal structure of Non-hydrolyzing UDPGlcNAc 2-epimerase | | Descriptor: | Putative UDP-N-acetylglucosamine 2-epimerase, SODIUM ION | | Authors: | Li, T.L, Rattinam, R. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | KasQ an Epimerase Primes the Biosynthesis of Aminoglycoside Antibiotic Kasugamycin and KasF/H Acetyltransferases Inactivate Its Activity.
Biomedicines, 10, 2022
|
|
7VZA
 
 | |
7VZ6
 
 | |
7AW0
 
 | | MerTK kinase domain in complex with purine inhibitor | | Descriptor: | 2-(cyclopentyloxy)-9-(2,6-difluorobenzyl)-N-methyl-9H-purin-6-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Clarke, M, Disch, J, Goldberg, K, Guilinger, J, Hennessy, E.J, Jetson, R, Ginkunja, D, Hardaker, E, Keefe, A, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S, Zhang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
6OWM
 
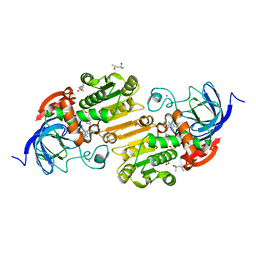 | | Horse liver F93W alcohol dehydrogenase complexed with NAD and pentafluorobenzyl alcohol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Substitutions of Amino Acid Residues in the Substrate Binding Site of Horse Liver Alcohol Dehydrogenase Have Small Effects on the Structures but Significantly Affect Catalysis of Hydrogen Transfer.
Biochemistry, 59, 2020
|
|
6OWP
 
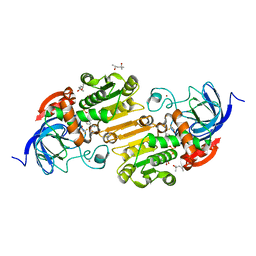 | | Horse liver F93W alcohol dehydrogenase complexed with NAD and trifluoroethanol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Substitutions of Amino Acid Residues in the Substrate Binding Site of Horse Liver Alcohol Dehydrogenase Have Small Effects on the Structures but Significantly Affect Catalysis of Hydrogen Transfer.
Biochemistry, 59, 2020
|
|
7AVY
 
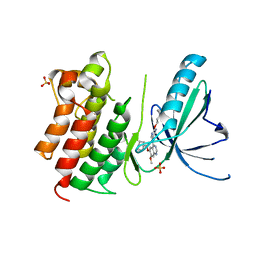 | | MerTK kinase domain in complex with quinazoline-based inhbitor | | Descriptor: | N-(2-(2-cyclopropylethoxy)pyrimidin-5-yl)-7-methoxy-6-(piperidin-4-ylmethoxy)quinazolin-4-amine, SULFATE ION, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AVZ
 
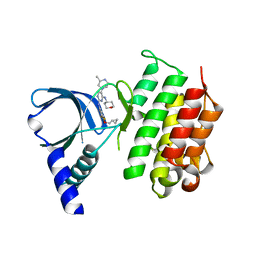 | | MerTK kinase domain in complex with a bisaminopyrimidine inhibitor | | Descriptor: | (R)-N2-(4-(cyclopropylmethoxy)-3,5-difluorophenyl)-5-(3-methylpiperazin-1-yl)-N4-(tetrahydro-2H-pyran-4-yl)pyrimidine-2,4-diamine, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AVX
 
 | | MerTK kinase domain in complex with NPS-1034 | | Descriptor: | 1-(4-fluorophenyl)-N-[3-fluoro-4-[(3-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)oxy]phenyl]-2,3-dimethyl-5-oxopyrazole-4-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW3
 
 | | MerTK kinase domain with type 1 inhibitor from a DNA-encoded library | | Descriptor: | 2-(1-((5-chloro-1H-pyrrolo[2,3-b]pyridine-3-carboxamido)methyl)-2-azabicyclo[2.1.1]hexan-2-yl)-N-methyl-4-(trifluoromethyl)thiazole-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW2
 
 | | MerTK kinase domain with type 1.5 inhibitor from a DNA-encoded library | | Descriptor: | 5-(2'-chloro-[1,1'-biphenyl]-4-yl)-N-(imidazo[1,2-a]pyridin-6-ylmethyl)-N-methyl-1,3,4-oxadiazol-2-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW4
 
 | | MerTK kinase domain with type 3 inhibitor from a DNA-encoded library | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-(4-methyl-1,2,3-thiadiazol-5-yl)-N-((R)-1-(((R)-3-(methylamino)-3-oxo-1-(4-(trifluoromethyl)phenyl)propyl)amino)-1-oxo-4-phenylbutan-2-yl)isoxazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW1
 
 | | MerTK kinase domain in complex with a type 2 inhibitor | | Descriptor: | N-(6-(4-(3-(4-((5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)methyl)-3-(trifluoromethyl)phenyl)ureido)phenoxy)pyrimidin-4-yl)cyclopropanecarboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7OH2
 
 | |
7OFX
 
 | | Crystal structure of a GH31 family sulfoquinovosidase mutant D455N from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | Alpha-glucosidase yihQ, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OFY
 
 | | Crystal structure of SQ binding protein from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | 1,2-ETHANEDIOL, Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Jarva, M.A, Sharma, M, Goddard-Borger, E.D, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OLF
 
 | |
