4X36
 
 | | Crystal structure of the autolysin LytA from Streptococcus pneumoniae TIGR4 | | Descriptor: | Autolysin, CHOLINE ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-11-28 | | Release date: | 2015-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Full-length structure of the major autolysin LytA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5WIN
 
 | | JAK2 Pseudokinase in complex with JNJ7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIJ
 
 | | JAK2 Pseudokinase in complex with NU6140 | | Descriptor: | 4-{[6-(cyclohexylmethoxy)-7H-purin-2-yl]amino}-N,N-diethylbenzamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIL
 
 | | JAK2 Pseudokinase in complex with AZD7762 | | Descriptor: | 5-(3-fluorophenyl)-N-[(3S)-3-piperidyl]-3-ureido-thiophene-2-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIK
 
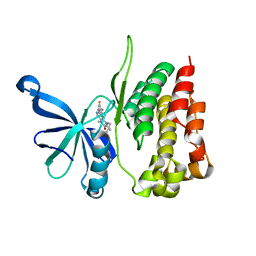 | | JAK2 Pseudokinase in complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIM
 
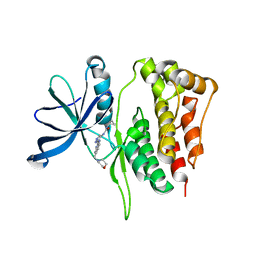 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
6BTD
 
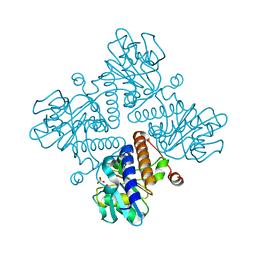 | |
6BTG
 
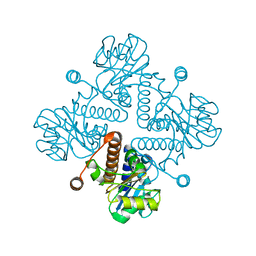 | |
6B9V
 
 | |
7H9O
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr13146 | | Descriptor: | Heat shock protein HSP 90-alpha, {4-[(oxan-4-yl)oxy]phenyl}methanol | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HB6
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr13755 | | Descriptor: | 5-fluoranyl-1~{H}-indole-2,3-dione, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBK
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr13232 | | Descriptor: | 6-(2,3-dimethylphenoxy)pyridin-3-amine, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HC1
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with 10T-0263 | | Descriptor: | (3M)-3-(3-fluoro-4-methoxyphenyl)-4-methyl-1H-pyrazole, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7H9V
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr12319 | | Descriptor: | 8-carbamoyl-1-benzopyran-1-ium, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7H9W
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr12314 | | Descriptor: | 3,4-dihydro-2~{H}-chromene-6-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HA0
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr12546 | | Descriptor: | 3-(pyridin-2-yloxy)aniline, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7H9R
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr14452 | | Descriptor: | Heat shock protein HSP 90-alpha, N-(3-ethynylphenyl)acetamide | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HA7
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with PS-5947 | | Descriptor: | 4-bromo-3-[(dimethylamino)methyl]phenol, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HAN
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with PS-3286 | | Descriptor: | (2P)-2-(1H-pyrazol-4-yl)pyrazine, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBR
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with AS-5576 | | Descriptor: | 4-(4-fluorophenyl)-1H-pyrazole, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7H9P
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr12541 | | Descriptor: | 4-phenoxyphenol, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7H9S
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with FS-3027 | | Descriptor: | Heat shock protein HSP 90-alpha, [2-(4-ethylpiperazin-1-yl)phenyl]methanol | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBM
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr13278 | | Descriptor: | Heat shock protein HSP 90-alpha, N-cycloheptyl-N'-(2-hydroxyethyl)thiourea | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBY
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with 9R-0342 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[(4-fluorophenyl)methyl]-1H-pyrrole-2-carboxamide | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7H9Y
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with NH-0224 | | Descriptor: | 1-[5-chloro-4-(difluoromethyl)pyridin-2-yl]piperazine, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
