8GFM
 
 | |
8GFQ
 
 | |
8GEZ
 
 | |
8GF0
 
 | |
8GFP
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with N-acetyl-2,3-dehydro-2-deoxyneuraminic acid inhibitor | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CITRIC ACID, Lytic transglycosylase domain-containing protein, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
8GFS
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with siastatin B inhibitor | | Descriptor: | (2S,3R,4S,5S)-2-(acetylamino)-5-carboxy-3,4-dihydroxypiperidinium, CITRIC ACID, Lytic transglycosylase domain-containing protein | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
8GFD
 
 | |
8GFI
 
 | |
8GFB
 
 | |
8GFL
 
 | |
1KSI
 
 | | CRYSTAL STRUCTURE OF A EUKARYOTIC (PEA SEEDLING) COPPER-CONTAINING AMINE OXIDASE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Wilce, M.C.J, Kumar, V, Freeman, H.C, Guss, J.M. | | Deposit date: | 1996-07-20 | | Release date: | 1997-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution.
Structure, 4, 1996
|
|
6VJE
 
 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
8SDV
 
 | | Crystal structure of PDC-3 Y221H beta-lactamase in complex with the boronic acid inhibitor S02030 | | Descriptor: | 1-{(2R)-2-(dihydroxyboranyl)-2-[(thiophen-2-ylacetyl)amino]ethyl}-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8SDN
 
 | | Crystal structure of PDC-3 Y221H beta-lactamase | | Descriptor: | Beta-lactamase, ISOPROPYL ALCOHOL | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8SDR
 
 | |
8SDT
 
 | |
8SDL
 
 | | Crystal structure of PDC-3 beta-lactamase | | Descriptor: | Beta-lactamase, IMIDAZOLE, ISOPROPYL ALCOHOL | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8SDS
 
 | |
4JQH
 
 | |
4RMP
 
 | | Crystal structure of allophycocyanin from marine cyanobacterium Phormidium sp. A09DM | | Descriptor: | Allophycocyanin, PHYCOCYANOBILIN | | Authors: | Kumar, V, Gupta, G.D, Sonani, R.R, Madamwar, D. | | Deposit date: | 2014-10-22 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal Structure of Allophycocyanin from Marine Cyanobacterium Phormidium sp. A09DM.
Plos One, 10, 2015
|
|
7LAM
 
 | | Crystal structure of Campylobacter jejuni Cj0843c lytic transglycosylase in complex with N,N',N''-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CITRIC ACID, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Turnover Chemistry and Structural Characterization of the Cj0843c Lytic Transglycosylase of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7LAQ
 
 | |
4O8B
 
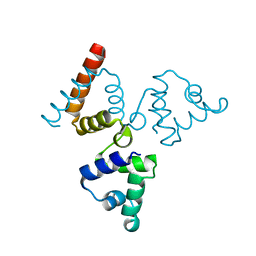 | | Crystal structure of transcriptional regulator BswR | | Descriptor: | Uncharacterized protein | | Authors: | Ye, F.Z, Wang, C, Kumar, V, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2013-12-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | BswR controls bacterial motility and biofilm formation in Pseudomonas aeruginosa through modulation of the small RNA rsmZ.
Nucleic Acids Res., 42, 2014
|
|
8RM5
 
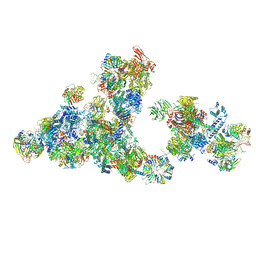 | | Cryo-EM structure of the cross-exon pre-B+5'ssLNG+ATPyS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'SS oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2024-01-05 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
4V90
 
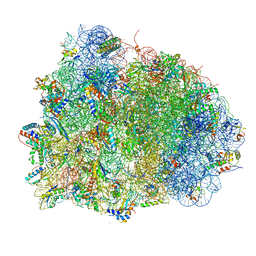 | | Thermus thermophilus Ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Chen, Y, Feng, S, Kumar, V, Ero, R, Gao, Y.G. | | Deposit date: | 2014-02-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of EF-G-ribosome complex in a pretranslocation state.
Nat. Struct. Mol. Biol., 20, 2013
|
|
