4GJ3
 
 | | Tyk2 (JH1) in complex with 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide | | Descriptor: | 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Ultsch, M.H. | | Deposit date: | 2012-08-09 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead Optimization of a 4-Aminopyridine Benzamide Scaffold To Identify Potent, Selective, and Orally Bioavailable TYK2 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4E4N
 
 | | JAK1 kinase (JH1 domain) in complex with compound 49 | | Descriptor: | Tyrosine-protein kinase JAK1, tert-butyl [(1R,3R)-3-(imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl)cyclopentyl]carbamate | | Authors: | Eigenbrot, C. | | Deposit date: | 2012-03-13 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4E6Q
 
 | | JAK2 kinase (JH1 domain) triple mutant in complex with compound 12 | | Descriptor: | 1-(1-benzylpiperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK2 | | Authors: | Murray, J.M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-30 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4EHZ
 
 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-1-(piperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4EI4
 
 | | JAK1 kinase (JH1 domain) in complex with compound 20 | | Descriptor: | (1R,3R)-3-(2-methylimidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(8H)-yl)cyclohexanol, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4F08
 
 | |
4F09
 
 | |
1NLD
 
 | | FAB FRAGMENT OF A NEUTRALIZING ANTIBODY DIRECTED AGAINST AN EPITOPE OF GP41 FROM HIV-1 | | Descriptor: | FAB1583 | | Authors: | Davies, C, Beauchamp, J.C, Emery, D, Rawas, A, Muirhead, H. | | Deposit date: | 1996-07-02 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Fab fragment from a neutralizing monoclonal antibody directed against an epitope of gp41 from HIV-1.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
4FK6
 
 | | JAK1 kinase (JH1 domain) in complex with compound 72 | | Descriptor: | N-({1-[(1R,2R,4S)-bicyclo[2.2.1]hept-2-yl]-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-2-yl}methyl)methanesulfonamide, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-06-12 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based discovery of C-2 substituted imidazo-pyrrolopyridine JAK1 inhibitors with improved selectivity over JAK2.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4QD6
 
 | | ITK kinase domain in complex with inhibitor compound | | Descriptor: | Tyrosine-protein kinase ITK/TSK, trans-4-({6-[(5-phenyl-1H-pyrazol-3-yl)amino]-4-(phenylsulfonyl)pyridin-2-yl}amino)cyclohexanol | | Authors: | McEwan, P.A, Barker, J.J, Eigenbrot, C. | | Deposit date: | 2014-05-13 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, synthesis and structure-activity relationships of a novel class of sulfonylpyridine inhibitors of Interleukin-2 inducible T-cell kinase (ITK).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8FFX
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 19980 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Reverse transcriptase/ribonuclease H, ... | | Authors: | Rumrill, S.R, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Targeting HIV-1 Reverse Transcriptase Using a Fragment-Based Approach.
Molecules, 28, 2023
|
|
7ZLG
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Mao, R, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLJ
 
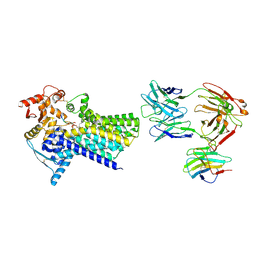 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLH
 
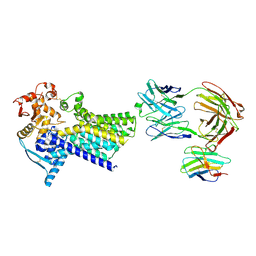 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLI
 
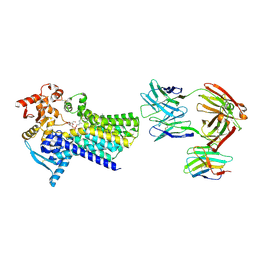 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
6N7B
 
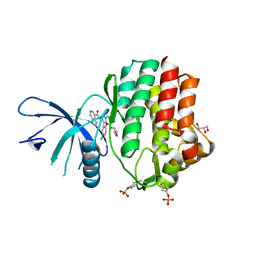 | | Structure of the human JAK1 kinase domain with compound 38 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrazolo[4,3-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N79
 
 | | Structure of the human JAK1 kinase domain with compound 20 | | Descriptor: | GLYCEROL, N-{5-[5-chloro-2-(difluoromethoxy)phenyl]-1H-pyrazol-4-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7C
 
 | | Structure of the human JAK1 kinase domain with compound 56 | | Descriptor: | GLYCEROL, N-[5-(3-methoxynaphthalen-2-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N77
 
 | | Structure of the human JAK1 kinase domain with compound 15 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7D
 
 | | Structure of the human JAK1 kinase domain with compound 54 | | Descriptor: | GLYCEROL, N-[5-(6-methoxy-1H-indazol-5-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N78
 
 | | Structure of the human JAK1 kinase domain with compound 21 | | Descriptor: | GLYCEROL, N-{3-[5-chloro-2-(difluoromethoxy)phenyl]-1-methyl-1H-pyrazol-4-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7A
 
 | | Structure of the human JAK1 kinase domain with compound 39 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]-2-methyl-2H-pyrazolo[4,3-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6NW2
 
 | | Structure of human RIPK1 kinase domain in complex with compound 11 | | Descriptor: | (5R)-5-methyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-4,5,6,7-tetrahydro-2H-indazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and selective inhibitors of receptor-interacting protein kinase 1 that lack an aromatic back pocket group.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1DEO
 
 | | RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.55 A RESOLUTION WITH SO4 IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RHAMNOGALACTURONAN ACETYLESTERASE, SULFATE ION, ... | | Authors: | Molgaard, A, Kauppinen, S, Larsen, S. | | Deposit date: | 1999-11-15 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases.
Structure Fold.Des., 8, 2000
|
|
1DEX
 
 | | RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.9 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RHAMNOGALACTURONAN ACETYLESTERASE, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Molgaard, A, Kauppinen, S, Larsen, S. | | Deposit date: | 1999-11-16 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases.
Structure Fold.Des., 8, 2000
|
|
