5MMH
 
 | |
6I09
 
 | |
6I0A
 
 | |
6I0N
 
 | |
5G5U
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa | | Descriptor: | BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-06-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G6T
 
 | | Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa | | Descriptor: | BETA-HEXOSAMINIDASE, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G1M
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, BETA-HEXOSAMINIDASE, CHLORIDE ION, ... | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
6I05
 
 | |
5G3R
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine and L-Ala-1,6-anhydroMurNAc | | Descriptor: | 2-[[(2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-oxidanyl-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoyl]amino]propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase, ... | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-04-30 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
6FCU
 
 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with 4(NAG-NAMpentapeptide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ALANINE, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FC4
 
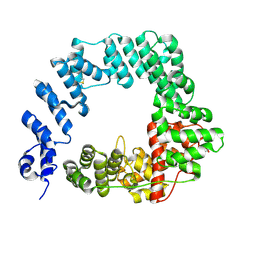 | |
6FCQ
 
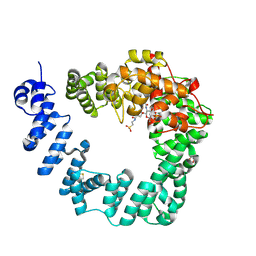 | |
6FCR
 
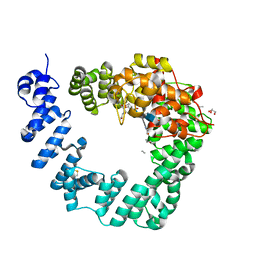 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMtetrapeptide-NAG-anhNAMtetrapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FCS
 
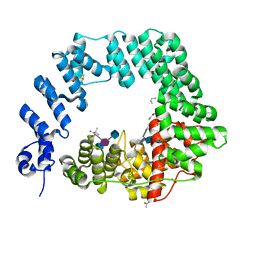 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMpentapeptide-NAG-NAMpentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBT
 
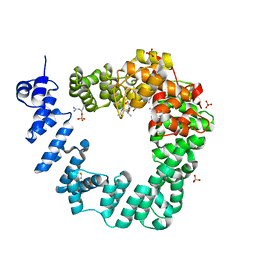 | | The X-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa in complex with the reaction product NAG-anhNAMpentapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, NAG-anhNAMpentapeptide, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4Y4V
 
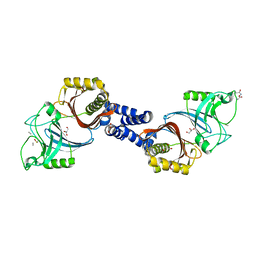 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
3TD5
 
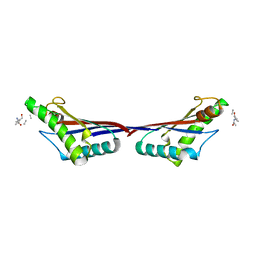 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, Outer membrane protein omp38, peptide(L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala) | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD4
 
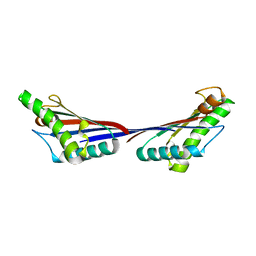 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
5AO7
 
 | | Crystal Structure of SltB3 from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Turnover of Bacterial Cell Wall by Sltb3, a Multidomain Lytic Transglycosylase of Pseudomonas Aeruginosa.
Acs Chem.Biol., 11, 2016
|
|
5AO8
 
 | |
5AA4
 
 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide | | Descriptor: | MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, [6-[[(2~{R})-1-azanyl-1-oxidanylidene-propan-2-yl]amino]-6-oxidanylidene-5-[[(4~{R})-5-oxidanyl-5-oxidanylidene-4-[[(2~{S})-2-[[(2~{R})-2-oxidanylpropanoyl]amino]propanoyl]amino]pentanoyl]amino]hexyl]azanium | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
5AA1
 
 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide | | Descriptor: | CHLORIDE ION, MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, N-ACETYLGLUCOSAMINE-1,6-ANHYDRO-N-ACETYLMURAMIC ACID L-ALA-D-GLU-M-DAP-D-ALA-D-ALA | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
5AA2
 
 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAM-pentapeptide. | | Descriptor: | CHLORIDE ION, MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, N-ACETYLGLUCOSAMINE-1,6-ANHYDRO-N-ACETYLMURAMIC ACID L-ALA-D-GLU-M-DAP-D-ALA-D-ALA | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
5AA3
 
 | |
5A5X
 
 | |
