5LOS
 
 | | Piriformospora indica PIIN_05872 | | Descriptor: | PIIN_05872, SULFATE ION, ZINC ION | | Authors: | Hartmann, M.D, Albrecht, R, Martin, J, Lupas, A.N. | | Deposit date: | 2016-08-09 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A secreted fungal histidine- and alanine-rich protein regulates metal ion homeostasis and oxidative stress.
New Phytol., 227, 2020
|
|
3ZMF
 
 | | Salmonella enterica SadA 303-358 fused to GCN4 adaptors (SadAK2) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PUTATIVE INNER MEMBRANE PROTEIN | | Authors: | Hartmann, M.D, Hernandez Alvarez, B, Albrecht, R, Lupas, A.N. | | Deposit date: | 2013-02-08 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A New Expression System for Protein Crystallization Using Trimeric Coiled-Coil Adaptors.
Protein Eng.Des.Sel., 21, 2008
|
|
2WG6
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A Mutant | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2XZR
 
 | | Escherichia coli Immunoglobulin-binding protein EibD 391-438 FUSED TO GCN4 ADAPTORS | | Descriptor: | CHLORIDE ION, IMMUNOGLOBULIN-BINDING PROTEIN EIBD | | Authors: | Hartmann, M.D, Hernandez Alvarez, B, Ridderbusch, O, Deiss, S, Lupas, A.N. | | Deposit date: | 2010-11-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Structure, 19, 2011
|
|
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WPY
 
 | | GCN4 leucine zipper mutant with one VxxNxxx motif coordinating chloride | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Zeth, K, Hartmann, M.D, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WPZ
 
 | | GCN4 leucine zipper mutant with two VxxNxxx motifs coordinating chloride | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Zeth, K, Hartmann, M.D, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4CTI
 
 | | Escherichia coli EnvZ histidine kinase catalytic part fused to Archaeoglobus fulgidus Af1503 HAMP domain | | Descriptor: | OSMOLARITY SENSOR PROTEIN ENVZ, AF1503 | | Authors: | Ferris, H.U, Coles, M, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2014-03-13 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Crystallographic Snapshot of the Escherichia Coli Envz Histidine Kinase in an Active Conformation.
J.Struct.Biol., 186, 2014
|
|
9QE5
 
 | | VCB in complex with VHL-binding compound 82 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-1-(1-fluoranylcyclopropyl)carbonylpiperidin-2-yl]carbonyl-~{N}-[(1~{S})-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Braun, M.B, Dierlamm, N, Hartmann, M.D. | | Deposit date: | 2025-03-07 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Breaking Barriers in PROTAC Design: Improving Solubility of USP7-Targeting Degraders
To Be Published
|
|
9QE4
 
 | | VCB in complex with VHL-binding compound 114 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Braun, M.B, Dierlamm, N, Hartmann, M.D. | | Deposit date: | 2025-03-07 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Breaking Barriers in PROTAC Design: Improving Solubility of USP7-Targeting Degraders
To Be Published
|
|
4USI
 
 | | Nitrogen regulatory protein PII from Chlamydomonas reinhardtii in complex with MgATP and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chellamuthu, V.R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Widespread Glutamine-Sensing Mechanism in the Plant Kingdom.
Cell(Cambridge,Mass.), 159, 2014
|
|
8BC6
 
 | |
8BC7
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex an aminoglutarimide degron peptide | | Descriptor: | Cereblon isoform 4, PHE-PHE-GLU-GLN-MET-GLN-QCI, S-Thalidomide, ... | | Authors: | Heim, C, Albrecht, R, Spring, A.K, Hartmann, M.D. | | Deposit date: | 2022-10-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Identification and structural basis of C-terminal cyclic imides as natural degrons for cereblon.
Biochem.Biophys.Res.Commun., 637, 2022
|
|
8RDR
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8g | | Descriptor: | (5~{S})-3-(2-nitrophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDS
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8i | | Descriptor: | (5~{S})-3-(2-methoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDT
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8j | | Descriptor: | (5~{S})-3-(2,3,4-trimethoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDQ
 
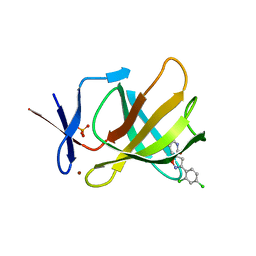 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8b | | Descriptor: | (5S)-3-(2,4-dichlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDP
 
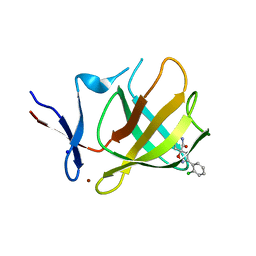 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8a | | Descriptor: | (5~{S})-3-(2-chlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
7R2Y
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in complex with ATP resulting from short ATP soak, conflicting T-loop and C-loop with partial occupancy | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R30
 
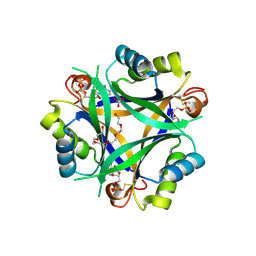 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in complex with ADP and AMP resulting from ADP soak | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R2Z
 
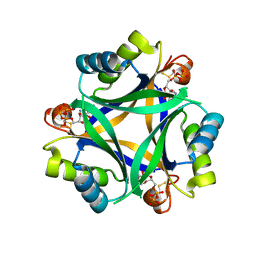 | |
7R32
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803, delta104 variant, in complex with ADP (co-crystal), tetragonal crystal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Membrane-associated protein slr1513 | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R31
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803, C105A+C110A variant, in complex with ATP (co-crystal), tetragonal crystal form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Membrane-associated protein slr1513, ... | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2VC8
 
 | |
8C3H
 
 | |
