7A0T
 
 | |
5E4M
 
 | |
5E4B
 
 | |
5E46
 
 | |
5E4D
 
 | |
5K3W
 
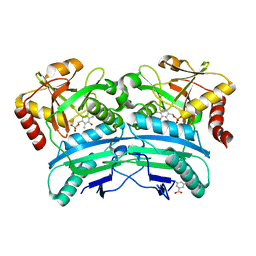 | | Structural characterisation of fold IV-transaminase, CpuTA1, from Curtobacterium pusillum | | Descriptor: | 3-AMINOBENZOIC ACID, CpuTA1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Pavkov-Keller, T, Diepold, M, Steiner, K, Gruber, K. | | Deposit date: | 2016-05-20 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Discovery and structural characterisation of new fold type IV-transaminases exemplify the diversity of this enzyme fold.
Sci Rep, 6, 2016
|
|
2XAA
 
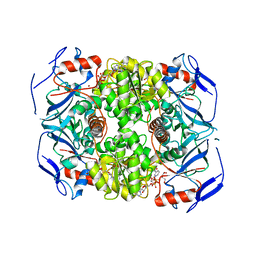 | | Alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541 at pH 8.5 in complex with NAD and butane-1,4-diol | | Descriptor: | 1,4-BUTANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SECONDARY ALCOHOL DEHYDROGENASE, ... | | Authors: | Kroutil, W, Gruber, K, Grogan, G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Substrate Specificity and Solvent Tolerance in Alcohol Dehydrogenase Adh-'A' from Rhodococcus Ruber Dsm 44541.
Chem.Commun.(Camb.), 46, 2010
|
|
4FQF
 
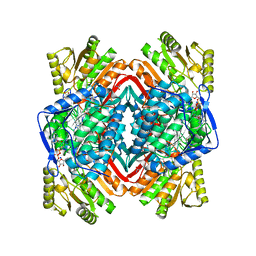 | |
4FR8
 
 | | Crystal structure of human aldehyde dehydrogenase-2 in complex with nitroglycerin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, ... | | Authors: | Lang, B.S, Gruber, K. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vascular Bioactivation of Nitroglycerin by Aldehyde Dehydrogenase-2: REACTION INTERMEDIATES REVEALED BY CRYSTALLOGRAPHY AND MASS SPECTROMETRY.
J.Biol.Chem., 287, 2012
|
|
4V15
 
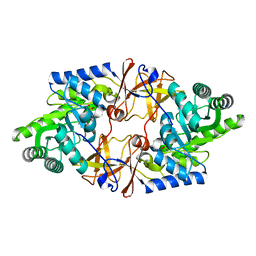 | | Crystal structure of D-threonine aldolase from Alcaligenes xylosoxidans | | Descriptor: | D-THREONINE ALDOLASE, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Uhl, M.K, Oberdorfer, G, Steinkellner, G, Riegler, L, Schuermann, M, Gruber, K. | | Deposit date: | 2014-09-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of D-Threonine Aldolase from Alcaligenes Xylosoxidans Provides Insight Into a Metal Ion Assisted Plp-Dependent Mechanism.
Plos One, 10, 2015
|
|
4WJL
 
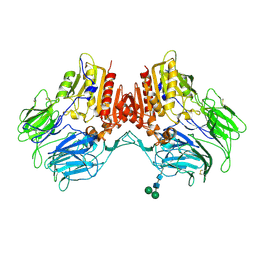 | | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive dipeptidyl peptidase 10, ... | | Authors: | Bezerra, G.A, Dobrovetsky, E, Seitova, A, Fedosyuk, S, Dhe-Paganon, S, Gruber, K. | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels.
Sci Rep, 5, 2015
|
|
5NY5
 
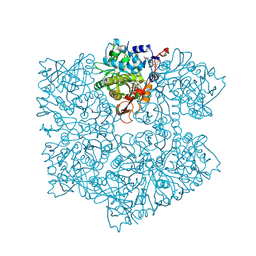 | | The apo structure of 3,4-dihydroxybenzoic acid decarboxylases from Enterobacter cloacae | | Descriptor: | 3,4-dihydroxybenzoate decarboxylase, GLYCEROL | | Authors: | Dordic, A, Gruber, K, Payer, S, Glueck, S, Pavkov-Keller, T, Marshall, S, Leys, D. | | Deposit date: | 2017-05-11 | | Release date: | 2017-09-13 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1JU2
 
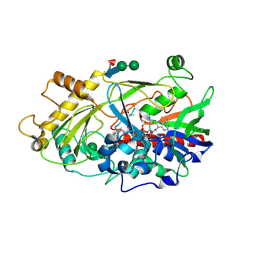 | | Crystal structure of the hydroxynitrile lyase from almond | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dreveny, I, Gruber, K, Glieder, A, Thompson, A, Kratky, C. | | Deposit date: | 2001-08-23 | | Release date: | 2002-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase.
Structure, 9, 2001
|
|
3RLI
 
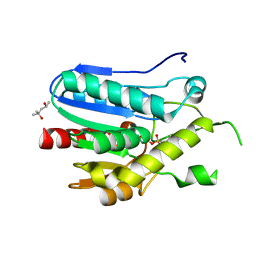 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 in complex with PMSF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase, phenylmethanesulfonic acid | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
3RM3
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
3FW7
 
 | |
3FWA
 
 | |
3FW8
 
 | |
3FW9
 
 | | Structure of berberine bridge enzyme in complex with (S)-scoulerine | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winkler, A, Macheroux, P, Gruber, K. | | Deposit date: | 2009-01-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural roles of biocovalent flaninylation in berberine bridge enzyme
to be published
|
|
6FRI
 
 | |
6GG2
 
 | | The structure of FsqB from Aspergillus fumigatus, a flavoenzyme of the amine oxidase family | | Descriptor: | Amino acid oxidase fmpA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pavkov-Keller, T, Lahham, M, Macheroux, P, Gruber, K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Oxidative cyclization ofN-methyl-dopa by a fungal flavoenzyme of the amine oxidase family.
J. Biol. Chem., 293, 2018
|
|
4CET
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with dicoumarol at 2.2 A resolution | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
4CF6
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with Cibacron blue at 2.7 A resolution | | Descriptor: | CIBACRON BLUE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
1CCW
 
 | |
4ZWN
 
 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | Descriptor: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | Authors: | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
